Mitophagy Detection Kit

Mitophagy Detection
- No need for Transfection
- Simply add Mtphagy Dye
-
Product codeMD01 Mitophagy Detection Kit
| Unit size | Price | Item Code |
|---|---|---|
| 1 set | Find your distributors | MD01-10 |
*Lyso Dye (inluded in MD01-10) stains lysosomes and is used to confirm the fusion of Mtphagy Dye-labeled mitochondria and lysosome. Therefore, we strongly recommend using Mitophagy Detection Kit (MD01) for first time users.
| 1 set | ・Mtphagy Dye ・Lyso Dye |
5 μg x 1 30 μg x 1 |
|---|
Description
Mitochondria is one of the cytoplasmic organelle that plays a crucial role in cells such as production of energy for cell viability. Recently, Mitophagy appears to be related to Alzheimer and Parkinson disease induced by the accumulation of depolarized mitochondria. Mitophagy serves as a specific elimination system that dysfunctional mitochondria caused by oxidative stress and DNA damage are sequestered into autophagosome, fused to lysosome and degraded by digestion.
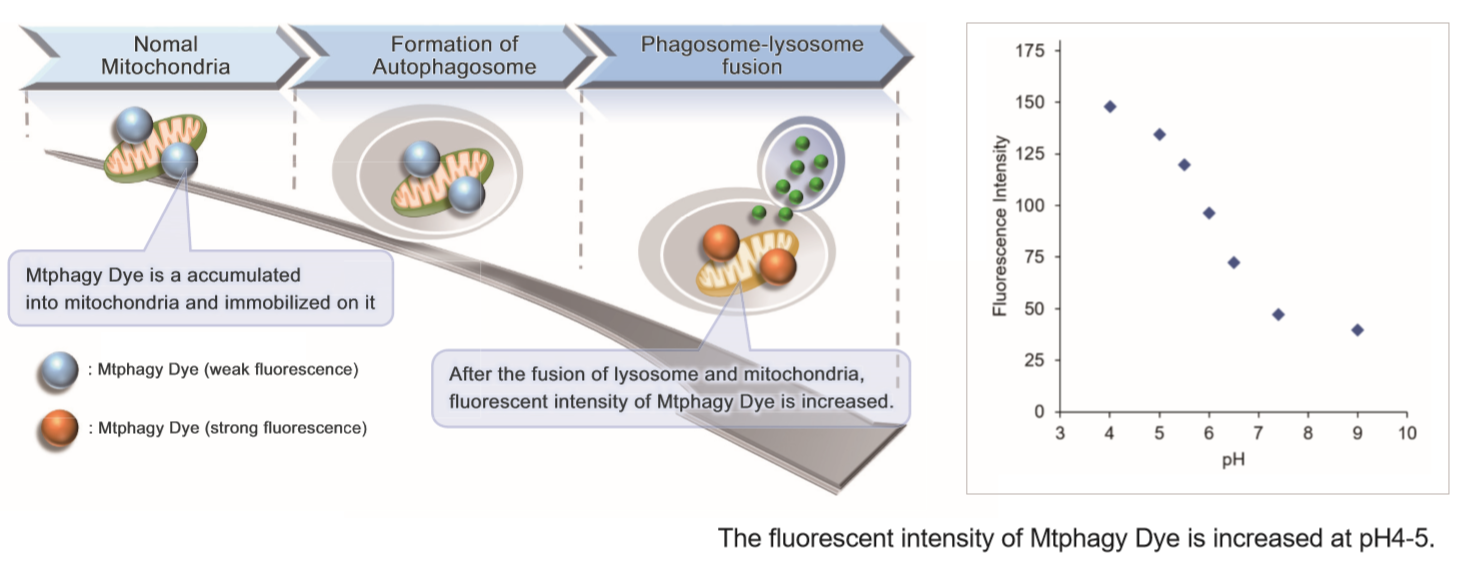
This kit is composed of Mtphagy Dye, reagent for detection of mitophagy, and Lyso Dye. Mtphagy Dye accumulates in intact mitochondria, is immobilized on it with chemical bond and exhibits a weak fluorescence from the influence of surrounding condition. When Mitophagy is induced, the damaged mitochondria fuses to lysosome and then Mtphagy Dye emits a high fluorescence. To confirm the fusion of Mtphagy Dye–labeled mitochondria and lysosome, Lyso Dye included in this kit can be used.
E. Fang etc. detected tomatidine induced mitophagy in HeLa cells by using mt-mKeima. Mitophagy was also detected in both primary rat cortical neurons and human SH-SY5Y neural cells using our Mitophagy Detection Kit.3 Mitophagy Detection Kit would be a valid alternative method if protein expression/transfection is not ideal for the experiment.
For more information on Mtphagy Dye compositions and examples, please refer to the publication below:
Iwashita H, Torii S, Nagahora N, Ishiyama M, Shioji K, Sasamoto K, Shimizu S, Okuma K. “Live Cell Imaging of Mitochondrial Autophagy with a Novel Fluorescent Small Molecule.“ACS Chem Biol, 2017, doi: 10.1021/acschembio.7b00647.
Notes: Mtphagy Dye and Lyso Dye are Patent Pending.
Mitophagy / Autophagy Analysis Products
| Product Name | Probe 1 Dyes and Fluorescence Properties |
Probe 2 Dyes and Fluorescence Properties |
|---|---|---|
| Mitophagy Detection Kit | Mtphagy Dye Ex: 500-560 nm, Em: 670-730 nm |
Lyso Dye Ex: 350-450 nm, Em: 500-560 nm |
| Mtphagy Dye | Mtphagy Dye Ex: 500-560 nm / Em: 690-750 nm |
|
| Autophagic Flux Assay Kit* | DAPRed Ex: 500-560 nm / Em: 690-750 nm |
DALGreen Ex: 350-450 nm / Em: 500-560 nm |
*Autophagic Flux Assay Kit includes lysosome acidification Inhibitor
Manual
Technical info
Transfection procedure is not needed for mitophagy imaging.
Experimental Example: Mitophagy Detection in Starved Cells
Living HeLa cells are co-stained with Mtphagy Dye and Lyso Dye under mitophagy induced condition.
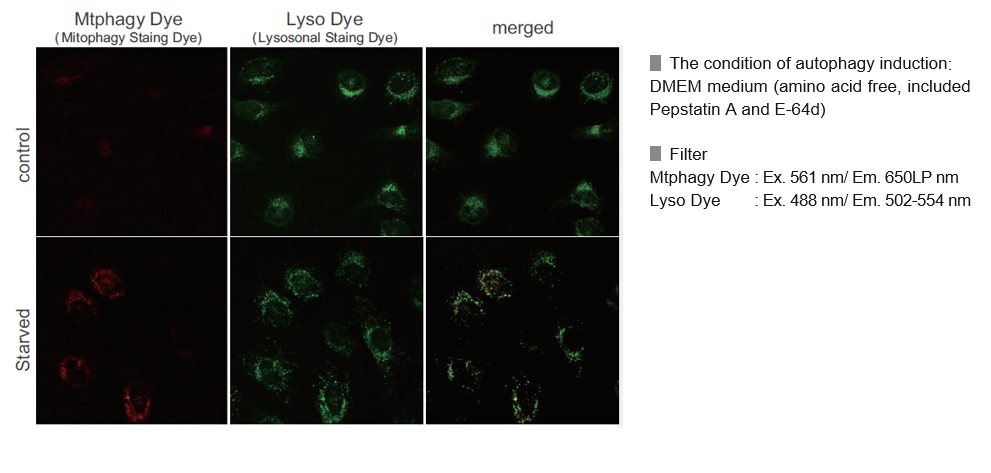
Mtphagy Dye was added to HeLa cells and the cells were incubated for 6 hours under starved condition. Before observation of the cells, lysosomal staining dye, Lyso Dye was added to HeLa cells. The fluorescent intensity of Mtphagy Dye was increased in the starved HeLa cells but not in normal cells. In addition, Mtphagy Dye was co-localized with Lyso Dye in the starved cells.
The amino acid-free medium used during starvation culture contained Pepstatin A 7.5 µmol/l and E-64d 1.0 µmol/l as final concentrations.
Experimental Example: Induction of Mitophagy in Parkin Expressed HeLa cells
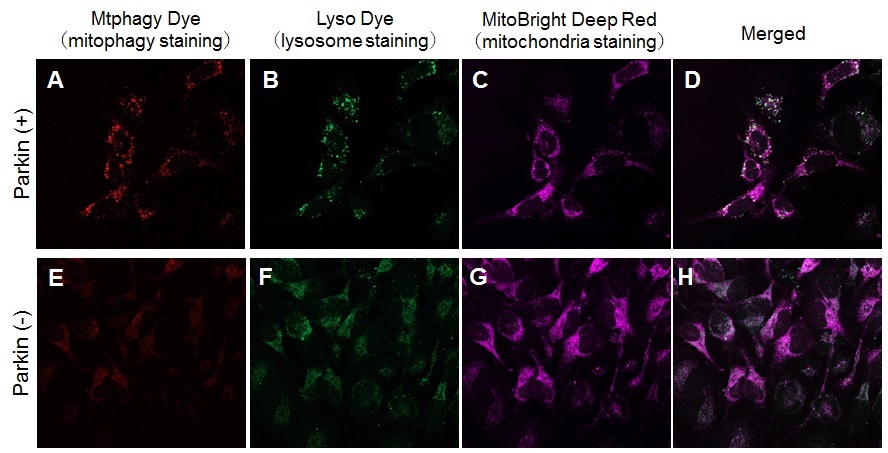
Induction of mitophagy by carbonyl cyanide m-chlorophenyl hydrazone (CCCP) as a mitochondrial-uncoupling reagent with Parkin expressed HeLa cells
HeLa cells were seeded on μ-slide 8 well (Ibidi) and cultured at 37oC overnight in a 5%-CO2 incubator. The cells were transfected with Parkin plasmid vector by HilyMax transfection reagent (Code#:H357), and incubated at 37oC overnight. The Parkin expressed HeLa cells were washed with Hanks’ HEPES buffer twice and then incubated at 37oC for 30 minutes with 250 μl of 100 nmol/l Mtphagy Dye working solution containing 100 nmol/l MitoBright LT Deep Red (※). After the washing of the cells with Hanks’ HEPES buffer twice, the culture medium containing 10 μmol/l CCCP was added to the well. After 24 hours incubation, mitophagy was observed by a fluorescence microscopy. After removing the supernatant, 250 μl of 1 μmol/l Lyso Dye working solution were added to the cells and incubated at 37oC for 30 minutes. The cells were washed with Hanks’ HEPES buffer twice and then co-localization of Mtphagy, Lyso Dye and MitoBright Deep Red was observed by confocal fluorescence microscopy.
Observation of mitophagy using Parkin
expressed HeLa cells (upper panel) and normal HeLa cells (Lower)
A, E) Fluorescent images of Mtphagy Dye; B, F) Fluorescent images of Lyso Dye;
C, G) Fluorescent images of MitoBright Deep Red (※); D, H) Merged images
※Our sincere apology that we discontinued distributing MitoBright Deep Red.
Please refer to MitoBright LT Deep Red (Code#:MT12) which improved retention ability.
Mitophagy Induction and Mitochondrial Membrane Potential Changes
Mitochondrial condition in the carbonyl cyanide m-chlorophenyl hydrazine (CCCP) treated Parkin-expressing HeLa cells was compared with untreated cells using Mitophagy Detection Kit (MD01, MT02) and JC-1 MitoMP Detection Kit (MT09).
Result: Mitophagy was hardly detected in the CCCP-untreated cells, and mitochondrial membrane potential was maintained normally. On the other hand, in CCCP-treated cells, we observed a decrease in mitochondrial membrane potential (decrease in red fluorescence of JC-1) and induction of mitophagy (increase in fluorescence of Mtphagy Dye).
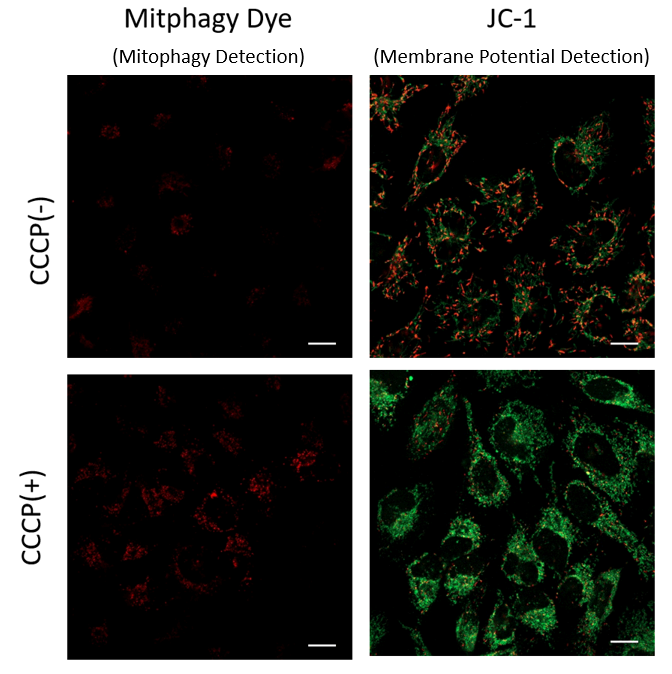
Transfection of Parkin plasmid to HeLa cells
- HileyMax (H357) was used to transfect Parkin plasmid to HeLa cells (Parkin plasmid/HilyMax reagent: 0.1μg/0.2 μL) by incubating overnight.
Detection of Mitophagy
1. Add 0.1 μmol/L Mtphagy working solution to Parkin expressing HeLa cells and incubated for 30 minutes at 37 ℃
2. Wash cells with HBSS
3. Add 10 μg/mL CCCP/MEM solution and inclubate for 2 hours at 37 ℃
4. Observe under fluorescence microscope
Detection of Mitochondrial Membrane Potential
1. Add 10 μg/mL CCCP/MEM solution to Parkin expressing HeLa cells and incubate for 1.5 hours at 37 ℃
2. Add 4 μmol/L JC-1 working solution (final concentration: 2 μmol/L) and incubate for 30 minutes at 37 ℃
3. Wash with HBSS and add Imaging Buffer Solution.
4. Observe under fluorescence microscope
Mitophagy Detection
Ex: 561 nm, Em: 570-700 nm
Mitochondrial Membrane Potential Detection
Green Ex: 488 nm, Em: 500-550 nm
Red Ex: 561 nm, Em: 560-610 nm
Comparison with Autophagy Marker
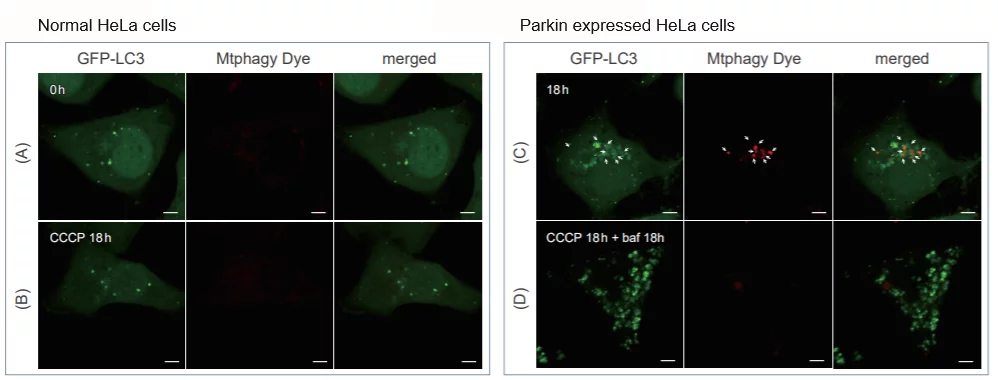
CCCP(carbonyl cyanide m-chlorophenyl hydrazone) has been added to normal and Parkin expressed cells. The strong fluorescence was not observed in normal HeLa cells(A)(B). On the other hand, the trong fluorescence was shown in Parkin expressed cells in 18 hours after additon of CCCP(C). Some of the puncta are co-localized with the autophagy marker(GFP-LC3). In addition, suppressed fluorescence of Mtphagy dye was observed when autophagy inhibitor, bafilomycin was added to Parking expressed cells(D). Because lysosomal pH was increased by the additon of bafilomycin.
Application
Flow Cytometer
Procedure for adherent cell
1. Cells were washed twice with DMEM and afterwards incubated at 37 °C for 30 min with 100 nmol Mtphagy Dye diluted in DMEM.
2. After this incubation cells were again washed twice with DMEM followed by the addition of complete DMEM.
3. The induction of mitophagy was then accomplished by the addition of 20 µM carbonyl cyanide 3-chlorophenylhydrazone (CCCP) for 24 h. Subsequently, fibroblasts were trypsinized and fluorescence intensity of Mtphagy Dye was measured by flow cytometry at 488 nm excitation and 655–730 nm emission.
Mitochondrial contribution to lipofuscin formation (Link to Reffered Publication)
Procedure for suspension cell
1. Briefly, 2×106 CD4+ T cells were divided into four tubes containing serum-free RPMI.
2. Tube #1 was for unstained cells that followed all washing and media changing processes.
3. 100 nmol/l Mtphagy Dye working solution was added to tubes #2, 3, 4 and then all tubes were incubated at 37°C for 30 minutes.
4. The cells were then washed with serum-free medium. After discarding the supernatant, complete medium (RPMI 1640, 10% FBS, 1% P/S/G) was added to all the tubes.
5. Ten μmol/l CCCP (Sigma-Aldrich) mitophagy-inducer was then added to tube #3, 100 nmol/l Bafilomycin A1 (Sigma-Aldrich) autophagy inhibitor was added to tube #4.
6. All tubes were then incubated at 37 °C for 18 hours.
7. After 18 hours, cells were washed with FACS buffer.
8. Mtphagy Dye fluorescence detection by flow cytometry (BD FACSCANTO II) was performed using 488 nm for excitation and 695 nm for emission (this corresponds to the PerCP cy5.5 channel). Data were analyzed using FLOWJO software (version 10).
Age-associated changes in human CD4+ T cells point to mitochondrial dysfunction consequent to impaired autophagy (Link to Reffered Publication)
Microplate Reader
References
1) J. Koniga, C. Otta, M. Hugoa, T. Junga, A. L. Bulteaub, T. Grunea and A. Hohna, "Mitochondrial contribution to lipofuscin formation", Redox Biology, 2017, 11, 673.
2) K. Kameyama, "Induction of mitophagy-mediated antitumor activity with folate-appended methyl-β-cyclodextrin", International Journal of Nanomedicine, 2017, 12, 3433.
3) E. F. Fang, T. B. Waltz, H. Kassahun, Q. Lu, J. S. Kerr, M. Morevati, E. M. Fivenson, B. N. Wollman, K. Marosi, M. A. Wilson, W. B. Iser, D. M. Eckley, Y. Zhang, E. Lehrmann, I. G. Goldberg, M. S. Knudsen, M. P. Mattson, H. Nilsen, V. A. Bohr and K. G. Becker, "Tomatidine enhances lifespan and healthspan in C. elegans through mitophagy induction via the SKN-1/Nrf2 pathway", Scientific Reports, 2017, 7, (46208), DOI: 10.1038/srep46208.
4) H. Iwashita, S. Torii, N. Nagahora, M. Ishiyama, K. Shioji, K. Sasamoto, S. Shimizu and K. Okuma, "Live Cell Imaging of Mitochondrial Autophagy with a Novel Fluorescent Small Molecule", ACS Chem. Biol., 2017, 12, (10), 2546.
5) Y. Feng, NB. Madungwe, CV. da Cruz Junho and JC. Bopassa, "Activation of G protein-coupled oestrogen receptor 1 at the onset of reperfusion protects the myocardium against ischemia/reperfusion injury by reducing mitochondrial dysfunction and mitophagy.", Br. J. Pharmacol., 2017, 174, (23), 4329.
6) K. M. Elamin, K. Motoyama, T. Higashi, Y. Yamashita, A. Tokuda and H. Arima, "Dual targeting system by supramolecular complex of folate-conjugated methyl-β-cyclodextrin with adamantane-grafted hyaluronic acid for the treatment of colorectal cancer.", Int. J. Biol. Macromol., 2018, doi: 10.1016/j.ijbiomac.2018.02.149.
7) N. Furuya, S. Kakuta, K. Sumiyoshi, M. Ando, R. Nonaka, A. Suzuki, S. Kazuno, S. Saiki and N. Hattori, "NDP52 interacts with mitochondrial RNA poly(A) polymerase to promote mitophagy.", EMBO Rep. ., 2018, doi: 10.15252/embr.201846363.
8) L. Zhu, X. Xie, L. Zhang, H. Wang, Z. Jie, X. Zhou, J. Shi, S. Zhao, B. Zhang, X. Cheng and S. Sun, "TBK-binding protein 1 regulates IL-15-induced autophagy and NKT cell survival", Nature Communications., 2018, 9, (1), doi:10.1038/s41467-018-05097-5.
9) K. Araki, K. Kawauchi, W. Sugimoto, D. Tsuda, H. Oda, R. Yoshida and K. Ohtani, "Mitochondrial protein E2F3d, a distinctive E2F3 product, mediates hypoxia-induced mitophagy in cancer cells", Commun Biol., 2019, DOI: 10.1038/s42003-018-0246-9.
10) E. Adegoke, S. Adeniran, Y. Zeng, X. Wang, H. Wang, C. Wang, H. Zhang, P. Zheng and G. Zhang , "Pharmacological inhibition of TLR4/NF-κB with TLR4-IN-C34 attenuated microcystin-leucine arginine toxicity in bovine Sertoli cells.", J Appl Toxicol., 2019,doi: 10.1002/jat.3771.
11) E. F. Fang, Y. Hou, K. Palikaras, B. A. Adriaanse, J. S. Kerr, B. Yang, S. Lautrup, M. M. Hasan-Olive, D. Caponio, X. Dan, P. Rocktaschel, D. L. Croteau, M. Akbari, N. H. Greig, T. Fladby, H. Nilsen, M. Z. Cader, M. P. Mattson, N. Tavernarakis and V. A. Bohr, "Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer's disease.", Nat. Neurosci. ., 2019,DOI:10.1038/s41593-018-0332-9.
12).Iwasawa, T. Shinomiya, N. Ota, N. Shibata, K. Nakata, I. Shiina, and Y. Nagahara , "Novel Ridaifen-B Structure Analog Induces Apoptosis and Autophagy Depending on Pyrrolidine Side Chain", Biological and Pharmaceutical Bulletin., 2019, 42, (3), 401-410, doi: 10.1248/bpb.b18-00643.
13) T. Namba, "BAP31 regulates mitochondrial function via interaction with Tom40 within ER-mitochondria contact sites ", Sci Adv., 2019, 5, (6), 1386.
14) A. Inamura, S. M. Hirayama, and K. Sakurai, Loss of Mitochondrial DNA by Gemcitabine Triggers Mitophagy and Cell Death', Biol. Pharm. Bull.., 2019, 42, 1977.
15) Y. Zhao and M. Sun, Metformin rescues Parkin protein expression and mitophagy in high glucose-challenged human renal epithelial cells by inhibiting NF-κB via PP2A activation., Life Sci.., 2020, DOI:10.1016/j.lfs.2020.117382.
16) N. Liu, J. Wu, L. Zhang, Z. Gao, Y. Sun, M. Yu, Y. Zhao, S. Dong, F. Lu and W. Zhang , "Hydrogen Sulphide modulating mitochondrial morphology to promote mitophagy in endothelial cells under high‐glucose and high‐palmitate ", J. Cell. Mol. Med., 2017, 21, (12), 3190.
17) N. Kajihara, D. Kukidome, K. Sada, H. Motoshima, N. Furukawa, T. Matsumura, T. Nishikawa and E. Araki, "Low glucose induces mitochondrial reactive oxygen species via fatty acid oxidation in bovine aortic endothelial cells", J Diabetes Investig, 2017, 8, (6), 750.
18) M. Jin, H. Ni and L. Li, "Leptin Maintained Zinc Homeostasis Against Glutamate-Induced Excitotoxicity by Preventing Mitophagy-Mediated Mitochondrial Activation in HT22 Hippocampal Neuronal Cells.", Front Neurol, 2018, 9, (9), 332.
19)D. Bhatia, K. P. Chung, K. Nakahira, E. Patino, M. C. Rice, L. K. Torres, T. Muthukumar, A. M. Choi, O. M. Akchurin and M. E. Choi , "Mitophagy-dependent macrophage reprogramming protects against kidney fibrosis", JCI Insight, 2019, 4, (23), e132826.
20) J. Zheng, D. L. Croteau, V. A. Bohr and M. Akbari, "Diminished OPA1 expression and impaired mitochondrial morphology and homeostasis in Aprataxin-deficient cells. ", Nucleic Acids Res., 2019, 47, (8), 4086.
21) D. D. Wang, M. F. Jin, D. J. Zhao and H. Ni, "Reduction of Mitophagy-Related Oxidative Stress and Preservation of Mitochondria Function Using Melatonin Therapy in an HT22 Hippocampal Neuronal Cell Model of Glutamate-Induced Excitotoxicity", Front Endocrinol (Lausanne), 2019, 10, 550.
22) A. Bektas, S. H. Schurman, M. G. Freire, A. Bektas, S. H. Schurman, M. G. Freire, C. A. Dunn, A. K. Singh, F. Macian, A. M. Cuervo, R. Sen and L. Ferrucci, "Age-associated changes in human CD4+ T cells point to mitochondrial dysfunction consequent to impaired autophagy.", Aging (Albany NY)., 2019, 11, (21), 9234-9263.
23) T. Nechiporuk, S.E. Kurtz, O. Nikolova, T. Liu, C.L. Jones, A. D. Alessandro, R. C. Hill, A. Almeida, S. K. Joshi, M. Rosenberg, C. E. Tognon, A. V. Danilov, B. J. Druker, B. H. Chang, S. K McWeeney and J. W. Tyner, "The TP53 Apoptotic Network Is a Primary Mediator of Resistance to BCL2 Inhibition in AML Cells.", Cancer Discov., 2019, 9, (7), 919.
24) Y. Zhang, R. Sun, X. Li and W. Fang, "Porcine Circovirus 2 Induction of ROS Is Responsible for Mitophagy in PK-15 Cells via Activation of Drp1 Phosphorylation", Viruses., 2020, 12, (3), 289.
25) Y. Huo, W. Chen, X. Zheng, J. Zhao, Q. Zhang, Y. Hou, Y. Cai, X. Lu and X. Jin , "The protective effect of EGF-activated ROS in human corneal epithelial cells by inducing mitochondrial autophagy via activation TRPM2.", J. Cell. Physiol., 2020, DOI: 10.1002/jcp.29597.
26) Y. Sun, F. Lu, X. Yu, B. Wang, J. Chen, F. Lu, S. Peng, X. Sun, M. Yu, H. Chen, Y. Wang, L. Zhang, N. Liu, H. Du, D. Zhao and W. Zhang, "Exogenous H2S Promoted USP8 Sulfhydration to Regulate Mitophagy in the Hearts of db/db Mice.", Aging Dis., 2020, 11, (2), 269.
27) R. Tanaka, M. Umemura, M. Narikawa, M. Hikichi, K. Osaw, T. Fujita, U. Yokoyama, T. Ishigami, K. Tamura and Y. Ishikawa, "Reactive fibrosis precedes doxorubicin-induced heart failure through sterile inflammation.", ESC Heart Fail., 2020, 7, (2), 588.
28) C. Duan, L. Kuang, X. Xiang, J. Zhang, Y. Zhu, Y. Wu, Q. Yan, L. Liu and T. Li, "Drp1 regulates mitochondrial dysfunction and dysregulated metabolism in ischemic injury via Clec16a-, BAX-, and GSH- pathways ", Cell Death Dis., 2020, 11, 251.
29) E. O. Adegoke, W. Xue, N. S. Machebe, S. O. Adeniran, W. Hao, W. Chen, Z. Han, Z. Guixue and Z. Peng, "Sodium Selenite inhibits mitophagy, downregulation and mislocalization of blood-testis barrier proteins of bovine Sertoli cell exposed to microcystin-leucine arginine (MC-LR) via TLR4/NF-kB and mitochondrial signaling pathways blockage.", Ecotoxicol. Environ. Saf., 2018, 116, 165.
30) D. Takahashi, J. Moriyama, T. Nakamura, E. Miki, E. Takahashi, A. Sato, T. Akaike, K. I. Nakama and H. Arimoto, "AUTACs: Cargo-Specific Degraders Using Selective Autophagy. ", Mol. Cell, 2019, 76, (5), 797.
31) H. Kim, J. H. Lee and J. W. Park, "IDH2 deficiency exacerbates acetaminophen hepatotoxicity in mice via mitochondrial dysfunction-induced apoptosis.", Biochim Biophys Acta Mol Basis Dis, 2019, 1865, (9), 2333.
32) M. S. Rahman and Y. S. Kim, "PINK1-PRKN mitophagy suppression by Mangiferin promotes a brown-fat-phenotype via PKA-p38 MAPK signalling in murine C3H10T1/2", Metabolism, 2020, 101, 154228.
33) W. Li, Y. Dai, B. Shi, F. Yue, J. Zou, G. Xu, X. Jiang, F Wang, X. Zhou and L. Liu, "LRPPRC sustains Yap-P27-mediated cell ploidy and P62-HDAC6-mediated autophagy maturation and suppresses genome instability and hepatocellular carcinomas ", Oncogene, 2020, 39, 3879.
34)S. Ikeoka and A. Kiso , "The Involvement of Mitophagy in the Prevention of UV-B-Induced Damage in Human Epidermal Keratinocytes ", J. Soc. Cosmet. Chem. Jpn., 2020, 54(3), 252.
Q & A
-
Q
What is an advantage of the kit in comparison to Keima-Red?
-
A
Our kit uses a small molecular fluorescent probe and allows detection of Mitophagy phenomenon without expressing fluorescent protein.
-
Q
Can I use Mitophagy Detection Kit with fixed cells?
-
A
No, the kit is suitable for live cells because Mtphagy Dye accumulates in intact mitochondria.
-
Q
Why it is not allowed to mix Mtphagy dye and Lyso dye together?
-
A
Since fluorescence of Lyso dye fades after long periods of staining, if both dyes are added together, Lyso dye does not emit enough fluorescence for double staining. Hence, Lyso dye is added right before the observation.
-
Q
How many times can I assay with Mitophagy Detection Kit?
-
A
For 96 wells plates format (100 ul/well), the kit is sufficient for 5 plates. For 35 mm dish format (2ml), the kit is sufficient for 25 assays.
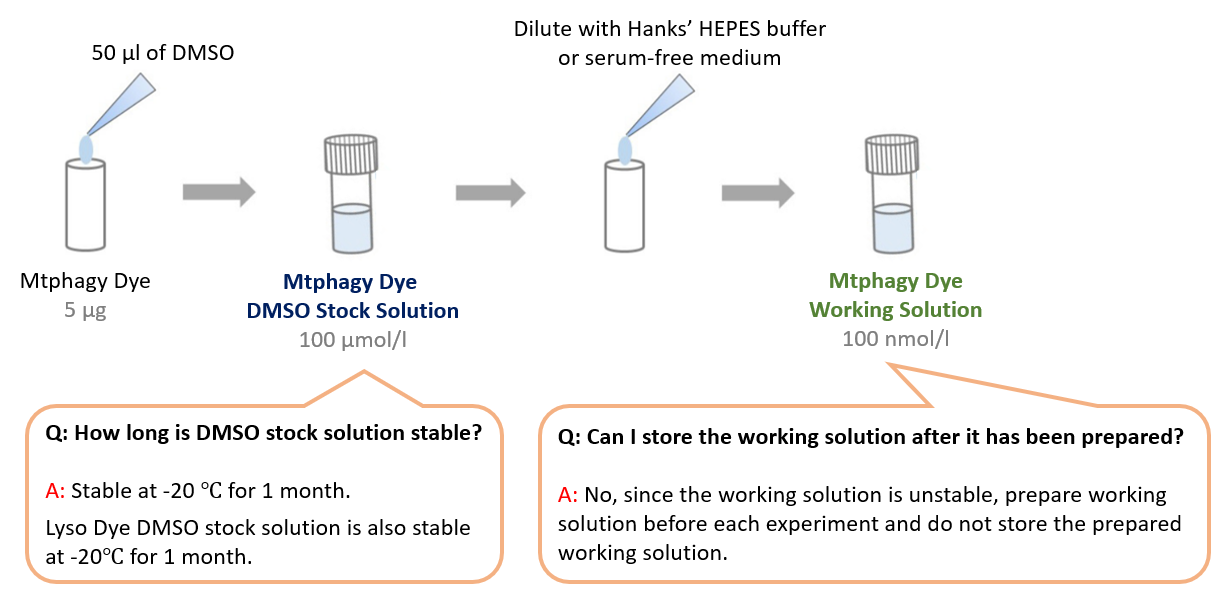
-
Q
What are some technical tips for successful assay?
-
A
1. Add Mtphagy Dye working solution BEFORE inducing mitophagy (Mtphagy Dye accumulates in the intact mitochondria)
2. Use appropriate filter – Mtphagy Dye: Ex.500-560 nm, Em.670-730 nm for microscopy detection
3. Use CCCP or FCCP for the positive control
*FCCP or CCCP concentration: 10 uM-30 uM, incubation time at 37℃ for 3 hours
FCCP: Carbonyl cyanide 4-(triguoromethoxy)phenylhydrazone
CCCP: Carbonyl cyanide 3-chlorophenylhydrazone
4. Use Confocal Microscope if available
-
Q
Is the serum in the medium susceptible to live cell imaging?
-
A
Yes, it is susceptible because both Mtphagy Dye and Lyso Dye interfere with the serum in the medium. Please use Hank’s HEPES buffer or serum-free medium.
-
Q
What is the recommended filter?
-
A
Mtphagy Dye: Ex.500~560 nm, Em.670~730 nm, Lyso Dye: Ex.350~450 nm, Em.500~560 nm
-
Q
Are there any points to be careful of Multi-Staining with MitoBright Deep Red?
-
A
The emission of Mtphagy Dye overlap with the emission of MitoBright Deep Red. You need to excite Mtphagy Dye at the wavelength that does not excite MitoBright Deep Red.
[Example] After staining MitoBright Deep Red, the cells were observed at the following excitation and emission filters for Mtpahgy dye and MitoBright Deep Red.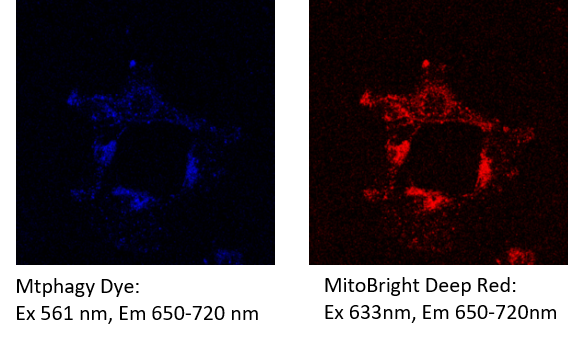
Fluorescence from the MitoBright Deep Red was detected in the channel of Mtphagy Dye.
Please refer to the following section to reduce spectral overlap to acceptable levels.
1. Change an excitation filterPlease change the excitation wavelength of Mtphagy Dye to approximately 500 nm and make sure that MitoBright Deep Red isn’t excited.
2. Adjust the excitation intensity and the sensitivity of fluorescent detectionPlease adjust the excitation intensity or the sensitivity of the detection until the fluorescent from the other dyes are no longer detected. After that, please double-check that you can detect the fluorescence of Mtphagy Dye.
How to check the spectral overlap
Example using Mtphagy Dye, Lyso Dye, and MitoBright Deep Red
1. Prepare cells in 3 different dishes or wells (dish/well No.1-3).
2. Add Mtphagy Dye to the dish/well No. 1, Add MitoBright Deep Red to dish/well No.2 with serum-free medium.
3. Incubate the cells at 37°C for 30 minutes.
4. Culture cells under the mitophagy inducing condition (e.g. starvation induced mitophagy)
5. Add Lyso Dye to No. 3 (No.3 is dish/well contained in only the cells) with serum-free medium.
6. Incubate the cells at 37°C for 30 minutes.
7. Detect fluorescence at the appropriate Ex./Em. wavelength for each reagent.
Mtphagy Dye: ex. 500-560nm, em. 670-730nm
Lyso dye: ex. 350-450nm, em. 500-560nm
MitoBright Deep Red: ex. 640nm, em. 656-700nm
8. Check to see if fluorescent is observed under detection condition of other reagents.(Example: For Dish/well No.1, please check to see the fluorescence under detection condition for Lyso dye and MitoBright Deep Red.)
-
Q
What is the staining mechanism of Lyso Dye?
-
A
Lyso Dye is a fluorescent dye that accumulates in normal lysosomes and then increases in intensity depending on the acidic conditions in the lysosomes.
The following products are available as fluorescent reagents with enhanced lysosomal retention.
[L261] LysoPrime Green - High Specificity and pH Resistance
-
Q
Can I use a fluorescence microscope to detect mitophagy?
-
A
It is possible to observe with a fluorescence microscope. However, we recommend using a confocal laser microscope to clearly image the mitophagy signals from the Mtphagy dye. When imaging with a fluorescence microscope, please note the following points.
1. Use a high magnification lens (40x or more).
2. Phenol red in the culture medium may cause a background. Please use phenol red-free medium or HBSS.
-
Q
Is the Mtphagy Dye compatible with flow cytometry?
-
A
Yes. Here is an example of an experiment using Mtphagy Dye in SH-SY5Y cells.
<Protocol>
1.SH-SY5Y cells(8×105 cells/well) were seeded in a 6 well plate and cultured overnight.
2.The Supernatant was removed, and the cells were washed serum-free medium .
3.100 nmol/l Mtphagy Dye working solution prepared in serum-free medium was added to a well and incubated for 30 minutes.
4.The supernatnat was removed and the cells were washed twice with serum-free medium.
5.Culture medium containing 10 µmol/l Oligomycin and 1 µmol/l Antimycin was added and incubated for 24 hours.
6.The supernatant was removed, and the cells were washed with PBS.
7.After trypsinization, 1 ml of the cell suspension was transferred to a microtube.
8.The cell suspension was centrifuged at 300×g for 5 minutes.
9.The supernatant was removed, and 1 ml of culture medium was added to the microtube and suspended the cells.
10.The cells were analyzed samples using a flow cytometer.
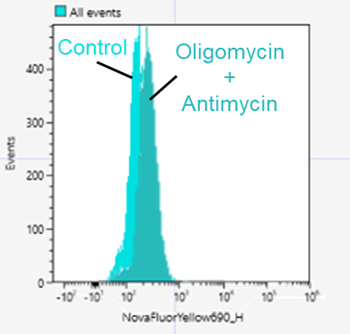
<Detection conditions>
Flow cytometer: SONY SA3800
Nova fluor Yellow 690(Ex:561nm, Em:655-735 nm)
*PE-AF610 or APC-Cy5.5 can be used as well.
Handling and storage condition
| 0-5°C, Protect from light, Nitrogen substitution, Protect from moisture |