DALGreen - Autophagy Detection

Autophagy (Autolysosome) Detection
- Allow to Monitor Autophagy at Real Time
- Detect Autolysosome selectively in Live Cells
- Very simple procedure. Just add the working solution reagent to cultured cells
-
Product codeD675 DALGreen - Autophagy Detection
| Unit size | Price | Item Code |
|---|---|---|
| 20 nmol | Find your distributors | D675-10 |
Product Description
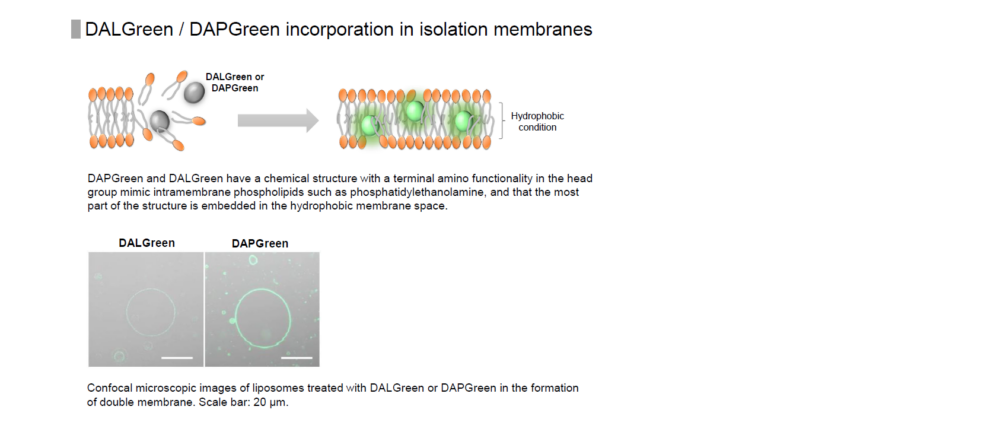
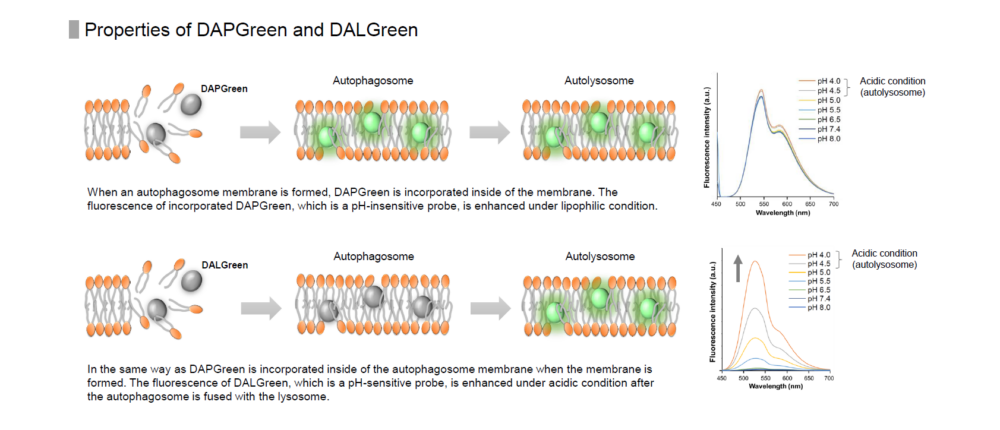
DALGreen is used to detect autophagy in live cells. Autophagy is an intracellular degradation system, where dysfunctional proteins and organelles are degraded. In this process, aggregated dysfunctional proteins are surrounded by the double membrane to form an autophagosome. DALGreen, which is a small hydrophobic molecule, passes the plasma membrane of live cells and is incorporated in the autophagosome. After a lysosome fuses with the autophagosome, the environment in the autolysosome become acidic. DALGreen fluoresce stronger as acidity increases. The quality of this dye enables live cell imaging with a fluorescence microscopy and quantitative assay by flow cytometry.
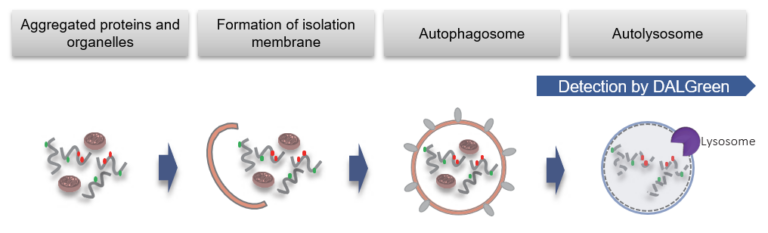
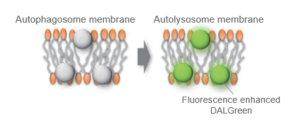
When an autophagosome membrane is formed, DALGreen is incorporated inside of the autophagosome membrane when the membrane is formed. The fluorescence of DALGreen is enhanced under acidic condition after the autophagosome is fused with the lysosome.
Autophagy Analysis Products
| Product Name | Autophagosome Detection Dyes and Fluorescence Properties |
Autolysosome Detection Dyes and Fluorescence Properties |
|---|---|---|
| Autophagic Flux Assay Kit* | DAPRed Ex: 500-560 nm / Em: 690-750 nm |
DALGreen Ex: 350-450 nm / Em: 500-560 nm |
| DAPGreen - Autophagy Detection | DAPGreen Ex: 425-475 nm / Em: 500-560 nm |
|
| DAPRed - Autophagy Detection | DAPRed Ex: 500-560 nm / Em: 690-750 nm |
|
| DALGreen - Autophagy Detection | DALGreen Ex: 350-450 nm / Em: 500-560 nm |
*Kit includes Lysosome Acidification Inhibitor
Manual
Technical info
First, cells are plated for the assay. After discarding the supernatant and washing the cells with culture medium, DALGreen working solution is added. While incubating for 30 minutes, DALGreen passes the plasma membrane of live cells and is incorporated in the forming autophagosome (Fig.1). After the 30 minutes incubation, supernatant is removed and the cells are washed. Medium containing autophagy-inducing agent is added to the cells. After incubating, observation can be made via fluorescence microscope or flow cytometer. For more detailed procedure, please refer to the manual.
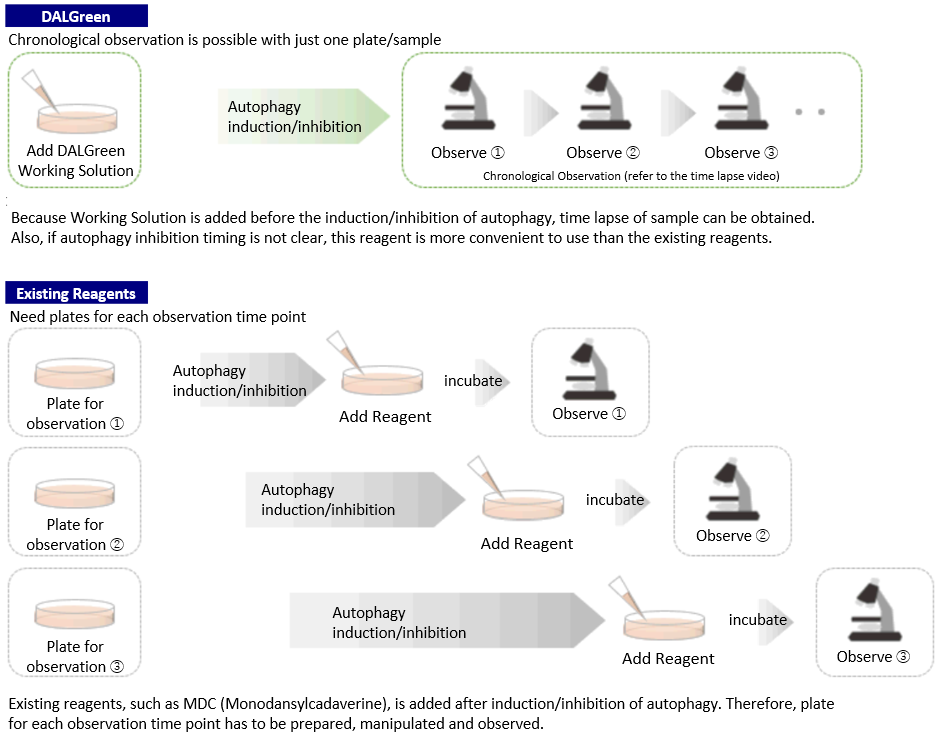
Procedure Comparison between DALGreen and Existing Reagents
Comparison to LC3
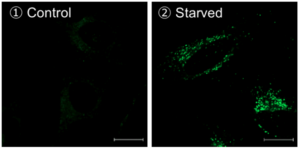
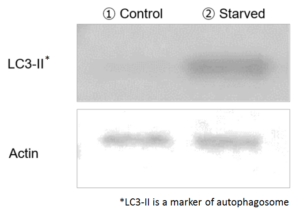
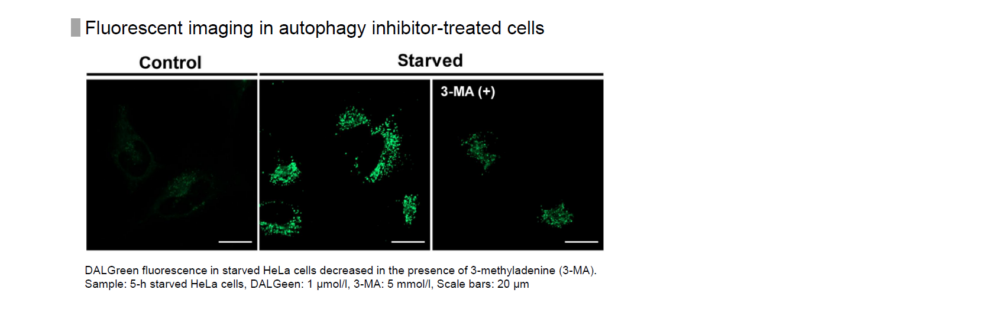
Method of autophagy induction
Control: incubated in the cell media for 6 hours
Starved: incubated in media without amino acid for 6 hours
Condition of DALGreen Imaging
Cell line: HeLa
Wavelength: Ex. 488 nm/ Em. 500-563 nm
Scale bar: 20 μm
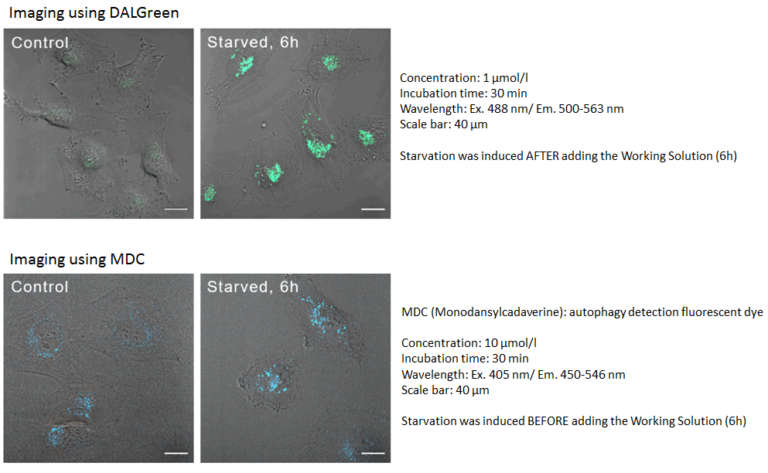
Comparison to MDC
DALGreen can be used for live autophagy monitoring because the Working Solution is added prior to inducing autophagy unlike MDC.
Reagent Properties
H. Iwashita, H. T. Sakurai, N. Nagahora, M. Ishiyama, K. Shioji, K. Sasamoto, K. Okuma, S. Shimizu, and Y. Ueno, “Small fluorescent molecules for monitoring autophagic flux“, FEBS Lett., 2018, 592, (4), 559–567.
Some data mentioned on this page is provided by the reference above.
Time-lapse imaging of autophagy
For more details, click application data
The Number of Usable Assay
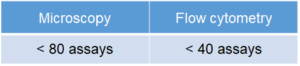
Microscopy: 250 μl /assay (8 well chamber slide)
Flow cytometry: 500 μl (24 well plate)
[Condition] ・Final concentration of DALGreen working solution: 1.0 μmol/l
・The total volume prepared at 1.0 μmol/l DALGreen working solution: 20 ml
NOTE: The number of assay depends on the final concentration of DALGreen or volume of working solution.
Usage example of publications
LC3 is one of markers that is used in autophagy detection1). However, whether or not autophagy is induced can’t be determined from only elevation of autophagy markers such as LC32). To increase the reliability of data, multiple assay approach is used in combination with western blot or different assay3).
Dojindo’s DALGreen, DAPGreen and DALRed can be used for the detection of fluorescent imaging. Their probes allows to visualize autophagosome/autolysosome selectively. DALGreen is used for autolysosome detection with western blot of LC3 4), 5).
1) I. Tanida, et al., “LC3 and Autophagy.” Methods Mol Biol., 2008, 445, 77-88.
2) DJ. Klionsky, et al., “Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition).”, Autophagy, 2016, 12(1), 1-222
3) S. Barth, et al., “Autophagy: assays and artifacts”, 2010, 221(2): 117–124.
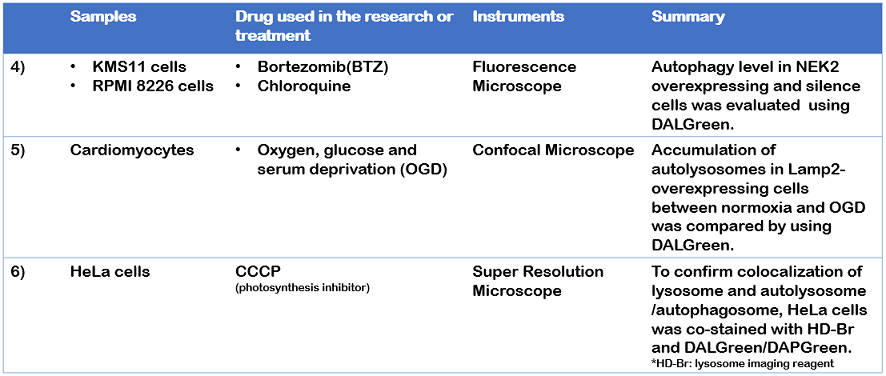
4) J. Xia, et al., “NEK2 Induces Autophagy-Mediated Bortezomib Resistance by Stabilizing Beclin-1 in Multiple Myeloma”, Mol Oncol., 2020, 14(4), 763-778.
5) L. Cui, et al., “The Lysosomal Membrane Protein Lamp2 Alleviates Lysosomal Cell Death by Promoting Autophagic Flux in Ischemic Cardiomyocytes”, Front. Cell Dev. Biol., 2020, 8, 31.
6) H. Fang et al., “De Novo-Designed Near-Infrared Nanoaggregates for Super-Resolution Monitoring of Lysosomes in Cells, in Whole Organoids, and in Vivo”, ACS Nano., 2019, 13(12), 14426-14436.
Related Product Information
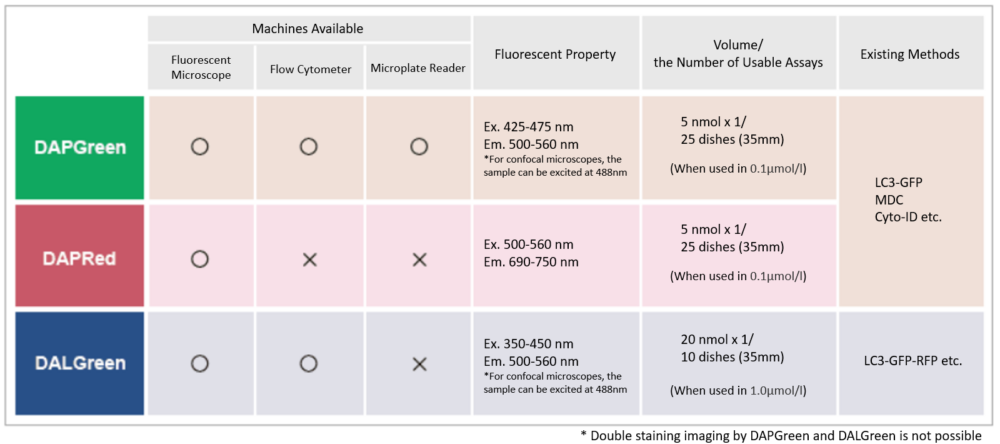
Detection is possible with a fluorescent microscope, a flow cytometer and a microplate reader using DAPGreen [#D676] and DAPRed [#D677]. DALGreen detects autolysosome. After a lysosome fuses with the autophagosome, the environment in the autolysosome become acidic. DALGreen fluoresce stronger as acidity increases. DALGreen can be applied in two methods (a fluorescent microscope and a flow cytometer). Please select your most suitable method depending on your equipment.
References
| No. | Sample | Instrument | Reference (Link) |
|---|---|---|---|
| 1) | Cell (HeLa, MEF) |
Fluorescence microscope | H. Iwashita, H. T. Sakurai, N. Nagahora, M. Ishiyama, K. Shioji, K. Sasamoto, K. Okuma, S. Shimizu, and Y. Ueno, "Small fluorescent molecules for monitoring autophagic flux.", FEBS Letters., 2018, 592, (4), 559–567. |
| 2) | Cell (HeLa) |
Fluorescence microscope | T. Sakata, A. Saito and H. Sugimoto, "In situ measurement of autophagy under nutrient starvation based on interfacial pH sensing.", Scientific Reports., 2018, 8, 8282. |
| 3) | Cell (HS-MM) |
Fluorescence microscope | Y. Egawa, C. Saigo, Y. Kito, T. Moriki and T. Takeuchi, "Therapeutic potential of CPI-613 for targeting tumorous mitochondrial energy metabolism and inhibiting autophagy in clear cell sarcoma.", PLoS One., 2018, 13, (6), e0198940. |
| 4) | Cell (HaCaT) |
Fluorescence microscope | S. Abe, S. Hirose, M. Nishitani, I. Yoshida, M. Tsukayama, A. Tsuji and K. Yuasa, "Citrus peel polymethoxyflavones, sudachitin and nobiletin, induce distinct cellular responses in human keratinocyte HaCaT cells.", Biosci. Biotechnol. Biochem. ., 2018, 82, (12), 1347. |
| 5) | Cell (KGN) |
Fluorescence microscope | W. Yuping, M. Congshun, Z. Huihui, Z. Yuxia, C. Zhenguo and W. Liping, "Alleviation of endoplasmic reticulum stress protects against cisplatin-induced ovarian damage.", Reprod. Biol. Endocrinol., 2018,doi: 10.1186/s12958-018-0404-4. |
| 6) | Cell (BmN) |
Fluorescence microscope | S. Xue, F. Mao, D. Hu, H. Yan, J. Lei, E. Obeng, Y. Zhou, Y. Quan, and W. Yu, "Acetylation of BmAtg8 inhibits starvation-induced autophagy initiation.", Mol. Cell Biochem., 2019,doi: 10.1007/s11010-019-03513-y. |
| 7) | Cell (HeLa) |
Fluorescence microscope | F. Hongbao, Y. Shankun, C. Qixin, L. Chunyan, C. Yuqi, G. Shanshan, B. Yang, T. Zhiqi, L. Z. Amanda, T. Takanori, C.Yuncong, G. Zijian, H. Weijiang and D. Jiajie, "De Novo-Designed Near-Infrared Nanoaggregates for Super-Resolution Monitoring of Lysosomes in Cells, in Whole Organoids, and in Vivo.", ACS Nano, 2019, 13, (12), 1446. |
| 8) | Cell (RT-7) |
Fluo Cytometer | E. Sasabe, A. Tomomura, N. Kitamura and T. Yamamoto, "Metal nanoparticles-induced activation of NLRP3 inflammasome in human oral keratinocytes is a possible mechanism of oral lichenoid lesions.", Toxicol In Vitro., 2020, 62, 104663. |
| 9) | Cell (Myeloma Cells) |
Fluorescence microscope | J. Xia, Y. He, B. Meng, S. Chen, J. Zhang, X. Wu, Y. Zhu, Y. Shen, X. Feng, Y. Guan, C. Kuang, J. Guo, Q. Lei, Y. Wu, G. An, G. Li, L. Qiu, F. Zhan and W. Zhou, "NEK2 induces autophagy-mediated bortezomib resistance by stabilizing Beclin-1 in multiple myeloma.", Mol Oncol, 2020, DOI: 10.1002/1878-0261.12641. |
| 10) | Cell (Human L2) |
Fluorescence microscope | Q. Xu, W. Shi, P. Lv, W. Meng, G. Mao, C. Gong, Y. Chen, Y. Wei, X. He, J. Zhao, H. Han, M. Sun and K. Xiao, "Critical role of caveolin-1 in aflatoxin B1-induced hepatotoxicity via the regulation of oxidation and autophagy.", Cell Death Dis., 2020, 11(1), 6. |
| 11) | Cell (Cardiomyocytes) |
Fluorescence microscope | L Cui, LP Zhao, JY Ye, L Yang, Y Huang, X.P. Jiang, Q. Zhang, JZ. Jia, DX. Zhang and Y. Huang, "The Lysosomal Membrane Protein Lamp2 Alleviates Lysosomal Cell Death by Promoting Autophagic Flux in Ischemic Cardiomyocytes.", Front Cell Dev Biol, 2020,DOI:10.3389/fcell.2020.00031. |
| 12) | Cell (IPEC-J2) |
Fluorescence microscope | Y Yang, J Huang, J Li, H Yang and Y. Yin, "The Effects of Butyric Acid on the Differentiation, Proliferation, Apoptosis, and Autophagy of IPEC-J2 Cells..", Curr. Mol. Med., 2020, 20(4), 307. |
| 13) | Cell (Fibroblasts, Kidney epithelial cells) | Fluorescence microscope | M. M. Ivanova, J. Dao, N. Kasaci, B. Adewale, J. Fikry and O. G. Alpan, "Rapid Clathrin-Mediated Uptake of Recombinant α-Gal-A to Lysosome Activates Autophagy", Biomolecules, 2020, 10(6), 837. |
| 14) |
Cell (NHEKs) |
Fluorescence microscope | S. Ikeoka and A. Kiso, "The Involvement of Mitophagy in the Prevention of UV-B-Induced Damage in Human Epidermal Keratinocytes ", J. Soc. Cosmet. Chem. Jpn., 2020, 54(3), 252. |
| 15) | Cell (HeLa) |
Fluorescence microscope(Super-resolution) | Q. Chen, M. Hao, L. Wang, L. Li, Y. Chen, X. Shao, Z. Tian, R. A. Pfuetzner, Q. Zhong, A. T. Brunger, J. Guan and J. Diao, "Prefused lysosomes cluster on autophagosomes regulated by VAMP8", Cell Death Dis., 2021, doi:10.1038/s41419-021-04243-0. |
| 16) | Cell (SH-SY5Y) |
Fluorescence microscope | Chang-ki Oh, Nima Dolatabadi, Piotr Cieplak, Maria T. Diaz-Meco, Jorge Moscat, John P. Nolan, Tomohiro Nakamura and Stuart A. Lipton, "S-Nitrosylation of p62 Inhibits Autophagic Flux to Promote α-Synuclein Secretion and Spread in Parkinson’s Disease and Lewy Body Dementia", J. Neurosci., 2022, doi:10.1523/JNEUROSCI.1508-21.2022. |
Q & A
-
Q
How stable is DALGreen working solution?
-
A
DALGreen working solution cannot be stored and should be used up on the day it is prepared.
-
Q
How stable is DALGreen DMSO stock solution?
-
A
The stock solution should be stored at -20°C in a light-shielded condition and is stable for 1 month after preparation.
For long storage, it is recommended to divide the solution into smaller portions.
-
Q
Do you have experience in knocking down and evaluating proteins related to autophagy?
-
A
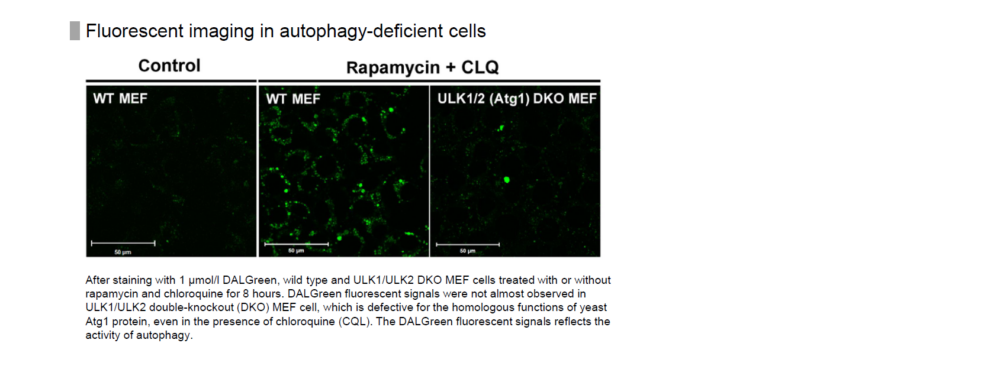
We have evaluated DALGreen in starvation of cells with knockdown of ULK1 and ULK2, which are involved in autophagosome membrane formation, as well as wild-type cells.
The result showed that the fluorescence of DALGreen increased only in wild-type cells not in ULK1/ULK2 knockdown cells.
Please refer to Supporting Information (Fig. S5) in the following article for this data.
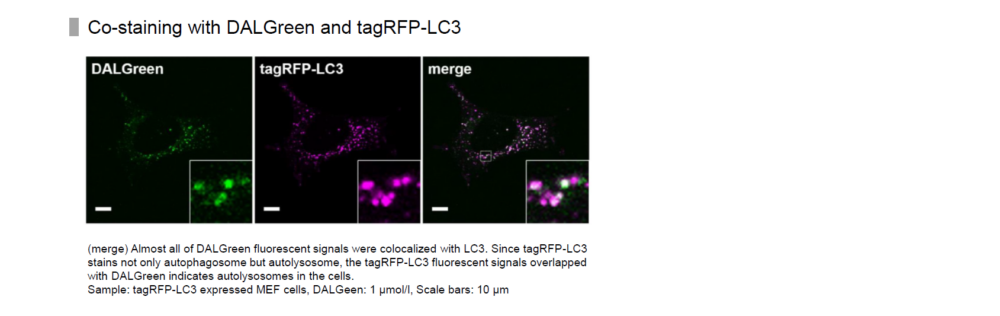
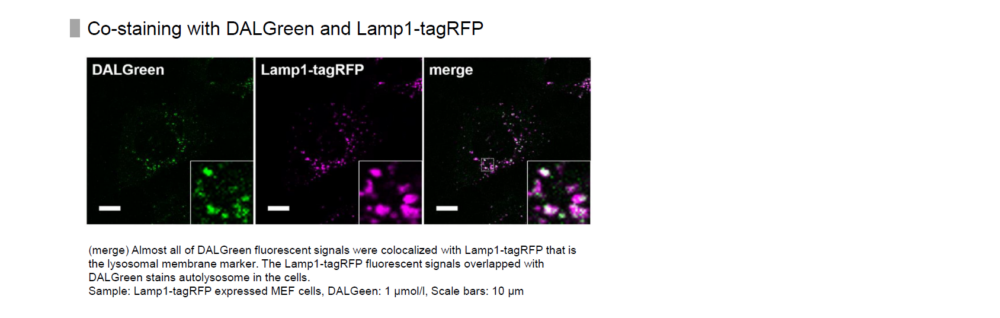
In addition, co-staining of LC3-RFP, a marker of autophagy, with DALGreen resulted in co-localization of most of the puncta. Please refer to Figure 1 in the following article.
H. Iwashita, H. T. Sakurai, N. Nagahora, M. Ishiyama, K. Shioji, K. Sasamoto, K. Okuma, S. Shimizu, and Y. Ueno, "Small fluorescent molecules for monitoring autophagic flux", FEBS Letters. 2018, 592, (4), 559-567.
-
Q
Is there any difference between this reagent and the lysosome staining reagent?
-
A
DALGreen localizes to autophagosomes and then its fluorescence increases when autophagosomes fuse with lysosomes.
When co-stained with lysosome staining reagent and DALGreen, the autolysosomal portion of the fluorescence derived from the lysosome staining reagent overlaps with that of DALGreen.
Please refer to Supporting Information (Fig. S5) in the following paper for this data.
H. Iwashita, H. T. Sakurai, N. Nagahora, M. Ishiyama, K. Shioji, K. Sasamoto, K. Okuma, S. Shimizu, and Y. Ueno, "Small fluorescent molecules for monitoring autophagic flux", FEBS Letters. 2018, 592, (4), 559-567.
-
Q
What excitation and emission conditions do you recommend?
-
A
Excitation: 350 - 450 nm
Emission: 500 - 560 nmWe have experience in fluorescence observation using a confocal microscope with 488 nm excitation.
Please refer to the examples of experiments on the product page of our website.
-
Q
How do I decide the optimal concentration of DALGreen?
-
A
If the reagent concentration is too high or too low, it may be difficult to tell the difference between autophagy induction and uninduced control.
It is recommended to consider the reagent concentration concerning the following information.The optimal concentration of DALGreen depends on the cell type.
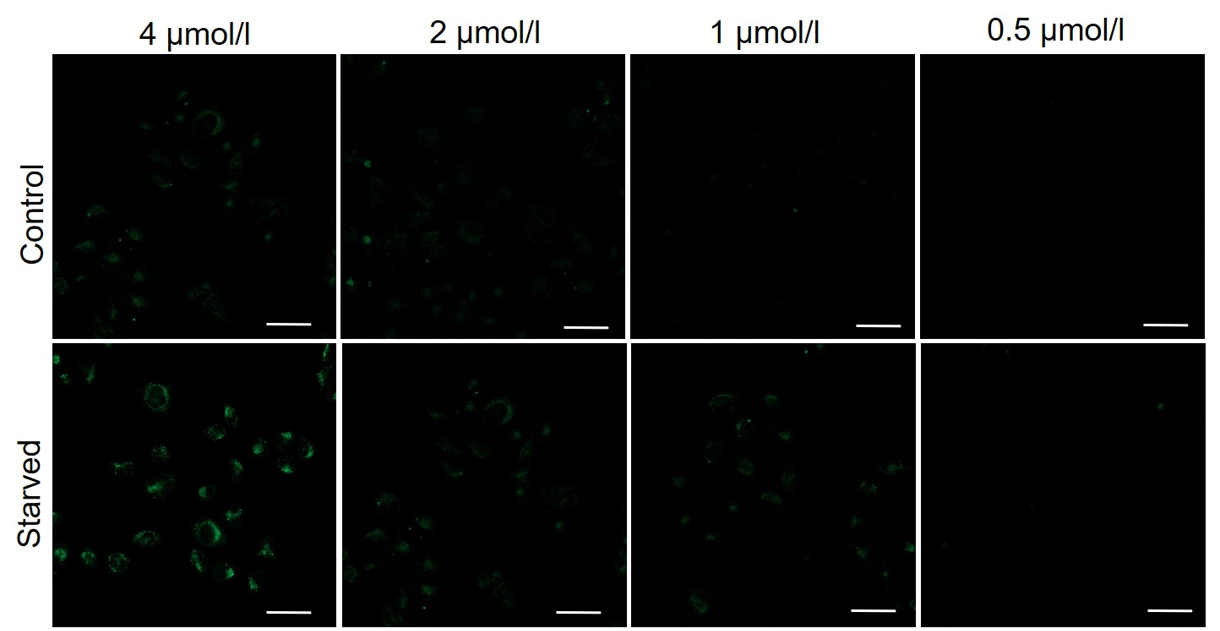
Please consider increasing the concentration of DALGreen in steps of several points starting from a low concentration (0.1 - 4 μmol/l is the approximate range).(Reference)
We have studied the optimal concentration of DALGreen for each cell type in HeLa, HepG2, and CHO cells.
DALGreen was stained at the following concentrations and cultured in an amino acid-free medium to induce autophagy.As a result, differences from the control were observed under the following concentration conditions highlighted in red.
Cell types Tested concentration HeLa 4 µmol/l, 2 µmol/l, 1 µmol/l, 0.5 µmol/l HepG2 4 µmol/l, 2 µmol/l, 1 µmol/l, 0.5 µmol/l CHO 4 µmol/l, 2 µmol/l, 1 µmol/l, 0.5 µmol/l HeLa
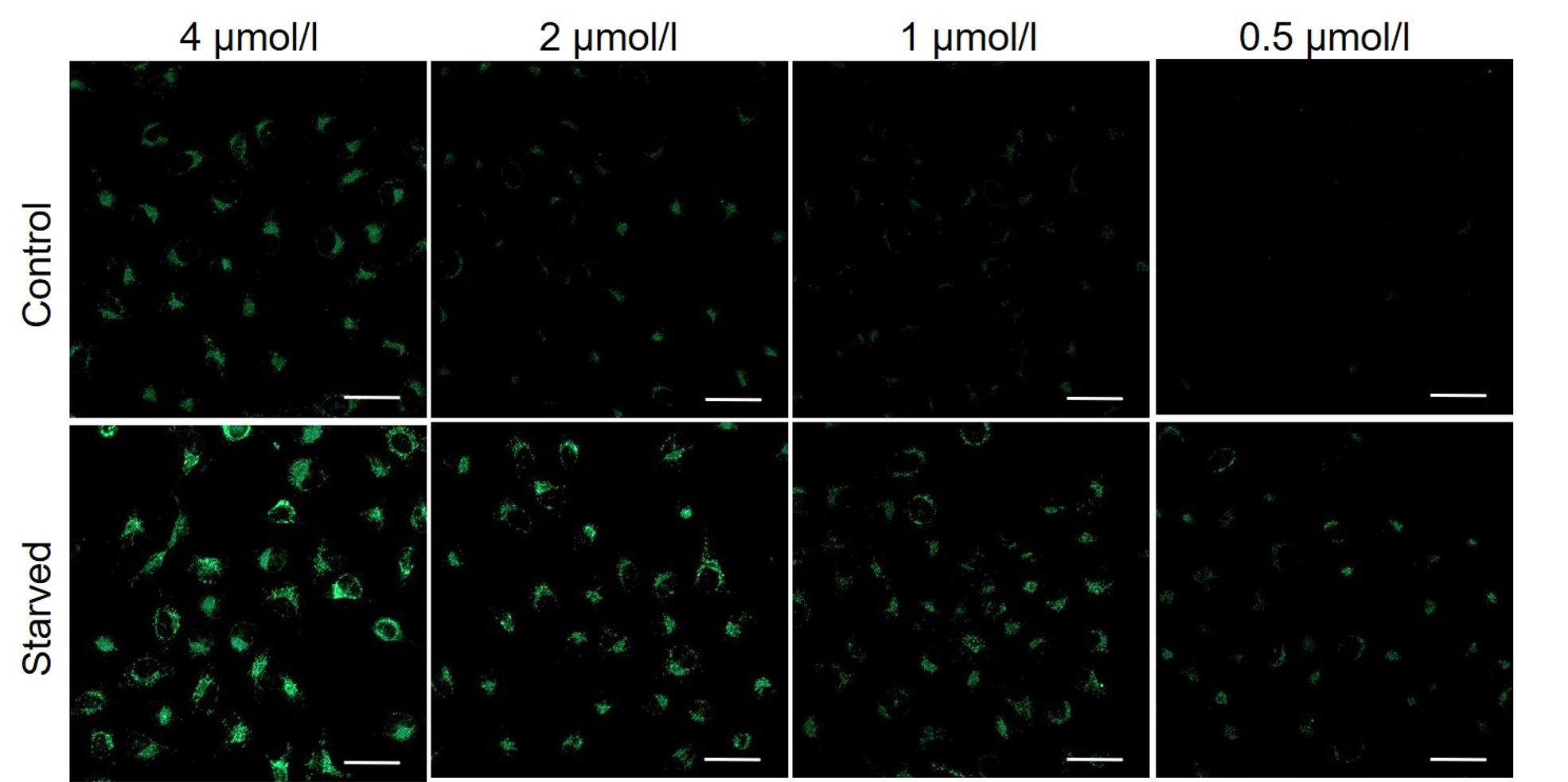
HepG2
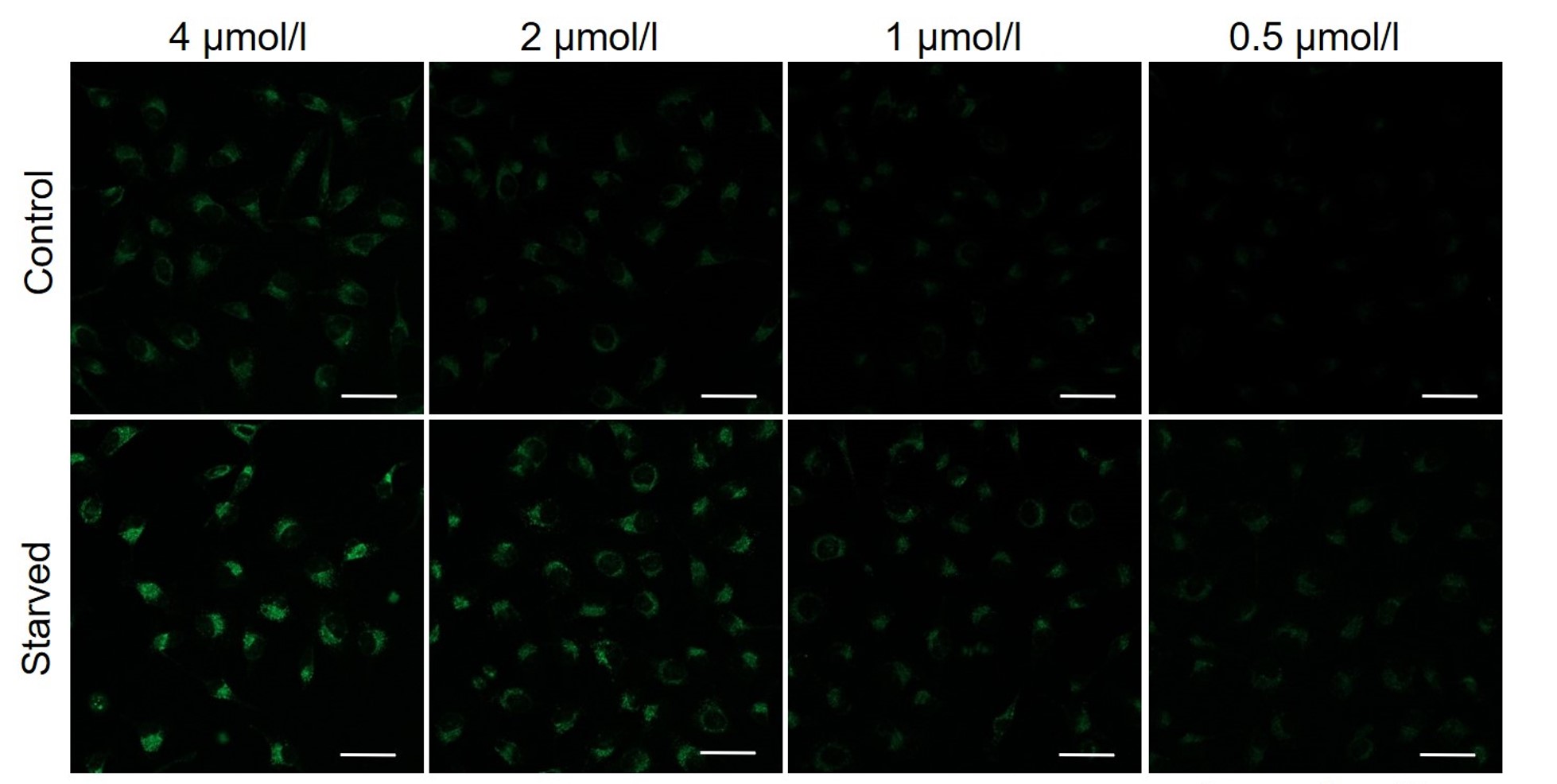
CHO
All Imaging Conditions:
DALGreen: Ex. 488 nm/Em. 500-563 nm
Scale bar: 50 μm
-
Q
Are there any precautions to take when performing time-lapse imaging?
-
A
Perform a preliminary experiment to set up the measurement conditions.
Due to the characteristics of the reagent, the initial fluorescence value tends to be high immediately after staining, so please perform preliminary experiments and time-lapse imaging by referring to the following procedures.(1) Preliminary experiment
・Use control cells (cells that do not induce autophagy).
・Stain cells with Working solution according to the instruction manual, and wash them twice with a culture medium.
・After adding a culture medium, observe the change in fluorescence over time.
・After staining, check the time when the fluorescence in the cells gradually decreases and then stabilizes (T in the figure).
Note: Conditions may vary depending on the cell type.(Reference)
In the case of HeLa cells, we have confirmed that the fluorescence intensity stabilizes after approximately 60 minutes of staining (DALGreen).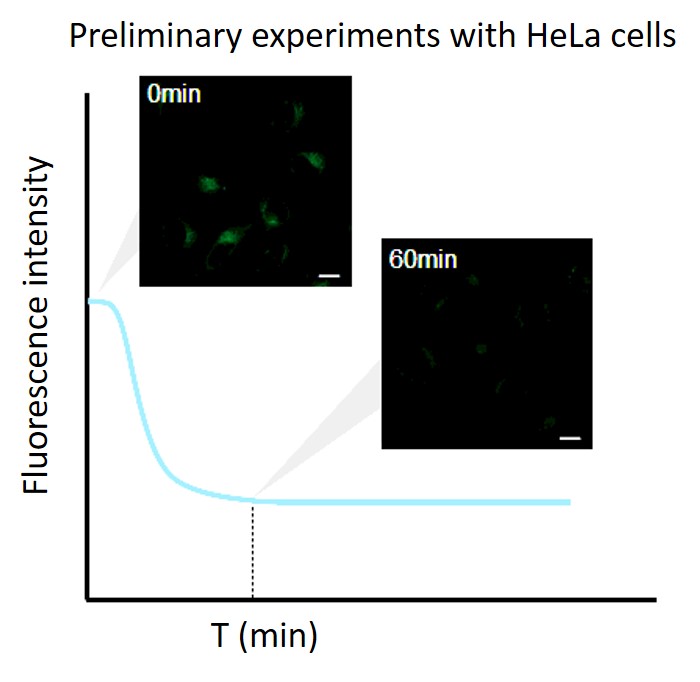
(2) Time-lapse imaging
・After staining cells with Working solution, incubate cells in a culture medium at 37°C.
Note 1: Incubate cells at 37°C in a culture medium for the time set in the preliminary experiment.
Note 2: Do not induce autophagy immediately after staining.・After incubation, start autophagy induction and perform time-lapse imaging.
(Reference)
HeLa cells were stained with DALGreen and incubated in a culture medium for 60 minutes (time set in the preliminary experiment) before autophagy induction.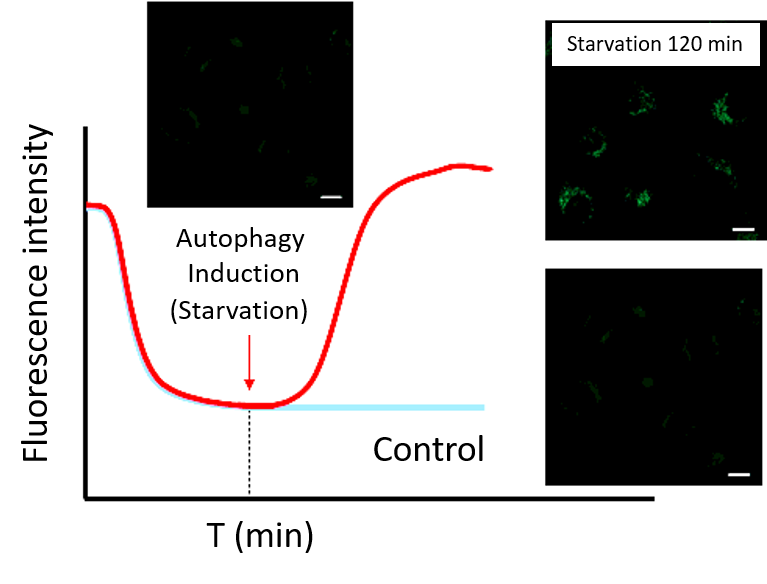
-
Q
How long can fluorescence be detected?
-
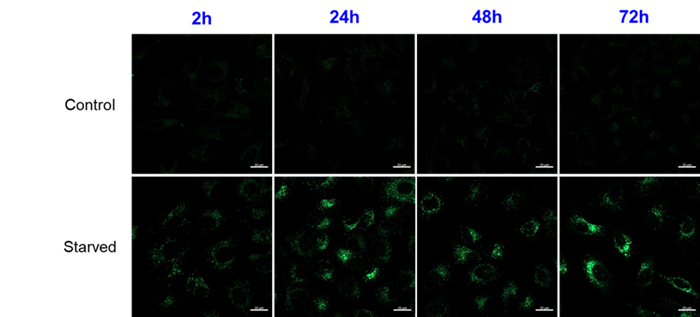
A
It can be observed for up to 72 hours after the staining.
Handling and storage condition
| Appearance: | Pale yellow solid |
|---|---|
| Purity (HPLC): | ≧ 90.0 % |
| -20°C, Protect from light, Nitrogen substitution |