|
Scientists have discovered that, in the absence of resident stem cells, senescent cells can instruct neighboring somatic cells to reprogram. This reprogramming enables them to become stem cells, driving whole-body regeneration in the cnidarian Hydractinia symbiolongicarpus. |
|||
|
Senescence-induced cellular reprogramming drives cnidarian whole-body regeneration |
|||
|
Point of Interest |
|||
| Related Techniques | |||
| First choice for cellular senescence assay | Cellular Senescence Detection Kit – SPiDER-ßGal | ||
| Cellular senescence assay with a plate reader | Cellular Senescence Plate Assay Kit – SPiDER-ßGal | ||
| Mitophagy detection | Mitophagy Detection Kit and Mtphagy Dye | ||
| Cell cycle assay | Cell Cycle Assay Solution Blue / Deep Red | ||
| Dead cell staining for flow cytometry | Dead Cell Makeup Blue / Deep Red - Higher Retention than PI | ||
| Lysosomal pH detection | Lysosomal Acidic pH Detection Kit-Green/Red, Green/Deep Red | ||
| Mitochondrial function/glycolysis detection | Glycolysis/JC-1 MitoMP Assay Kit | ||
| Oxygen consumption rate assay | Extracellular OCR Plate Assay Kit | ||
| Related Applications | |||
| Regulating Cell Cycle Arrest: p16, p21, p53, and pRB | |||
|
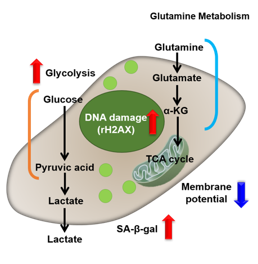
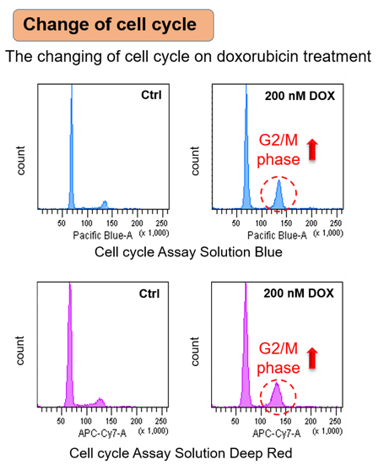
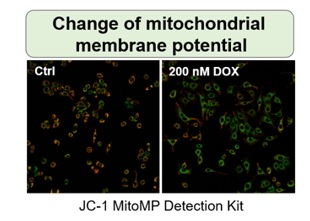
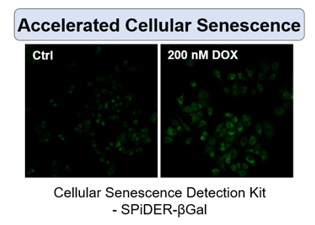
Irreversible cell cycle arrest is one of the phenomena that characterize cellular senescence. p16, p21, p53, and pRB (phosphorylated retinoblastoma protein) are known as representative protein markers. The activation/upregulation of these proteins are used as indicators of cellular senescence. These marker proteins are known to be tumor suppressors and regulate the cell cycle mainly through two pathways (p16Ink4a-RB and p53-p21CIP1).
|
||















