Science Note
New Mitophagy Pathways: Next Therapy Targets? [Sep. 23, 2025]
|
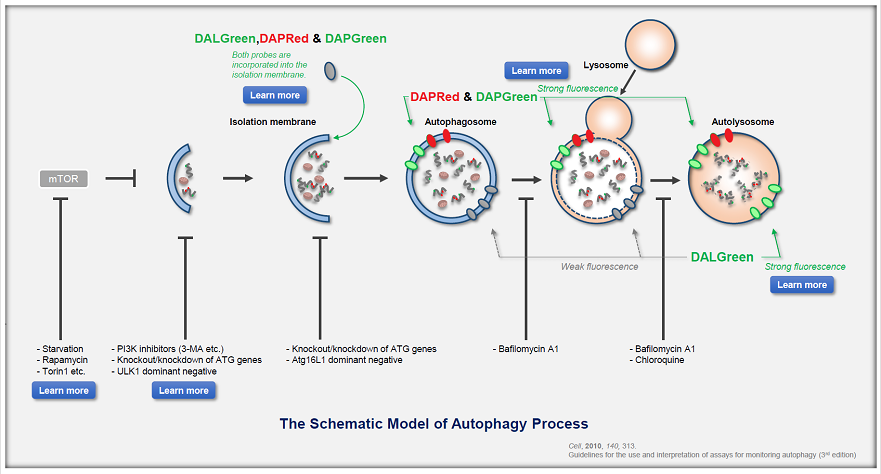
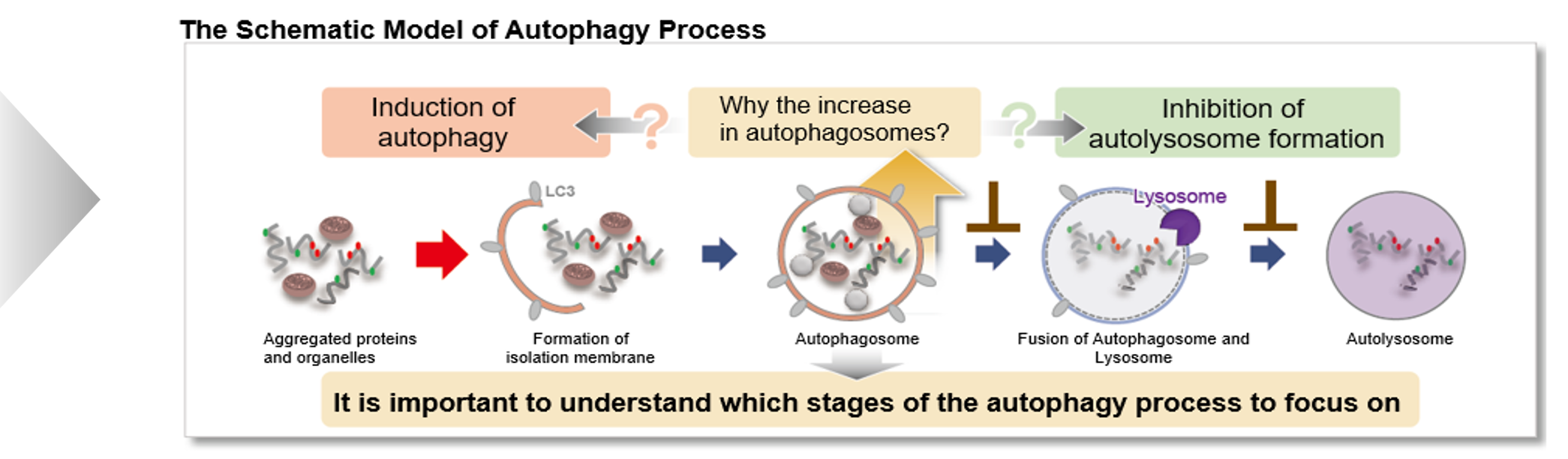
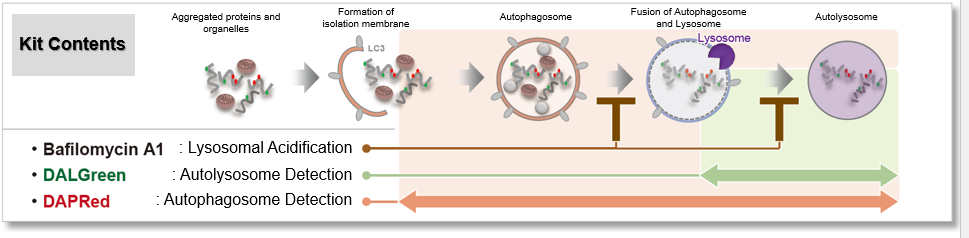
Defects in mitophagy through the autophagy–lysosome pathway, particularly in the PINK1/Parkin system, are closely linked to neurodegenerative disease. Recent studies reveal additional mechanisms, including a mitophagic stress response that relieves autophagy inhibition and a PINK1–SMAD3 transcriptional loop, highlighting new therapeutic targets in mitochondrial quality control. |
||||||||||||||||||||||
|
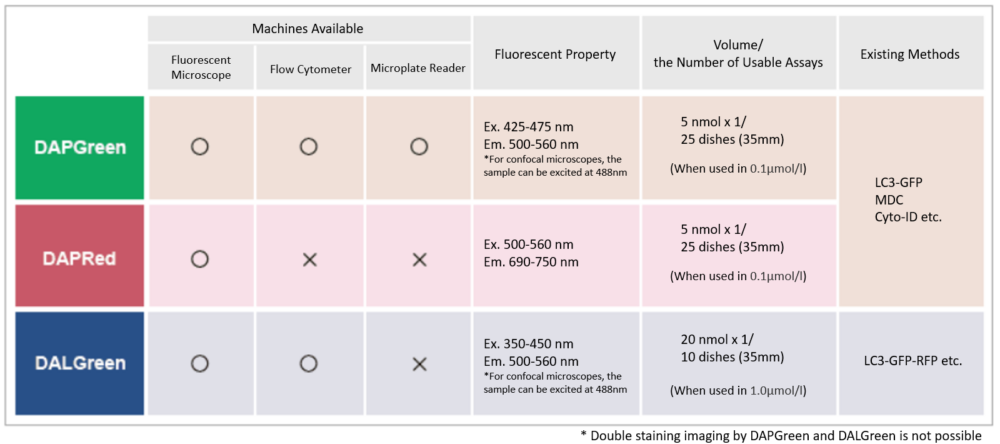
1. Mitochondrial damage triggers the concerted degradation of negative regulators of neuronal autophagy (Nature Communications, 2025) Highlighted technique: The study used fluorescent pH probes to monitor lysosomal acidity under mitochondrial stress. With this approach, the authors showed that reducing Rubicon, a protein that suppresses autophagy, restored lysosomal activity and enabled more efficient clearance of damaged mitochondria. Related techniques Lysosomal Function Analysis (as used in this article), Autophagosome Detection (as used in this article) |
||||||||||||||||||||||
|
2. A positive feedback loop between SMAD3 and PINK1 in regulation of mitophagy (Cell Discovery, 2025) Highlighted technique: To evaluate mitophagy, researchers used the Mito-Keima assay, which tracks mitochondria entering acidic lysosomes as a measure of degradation. In HeLa cells expressing YFP-Parkin and mito-Keima, SMAD3 knockdown combined with Oligomycin/Antimycin A treatment enabled flow cytometric quantification of this process. Related techniques Mitophagy Detection, Autophagic Flux Assay Kit |
||||||||||||||||||||||
Related Techniques (click to open/close)
|
||||||||||||||||||||||
Application Note I (click to open/close)
|
||||||||||||||||||||||
|
|
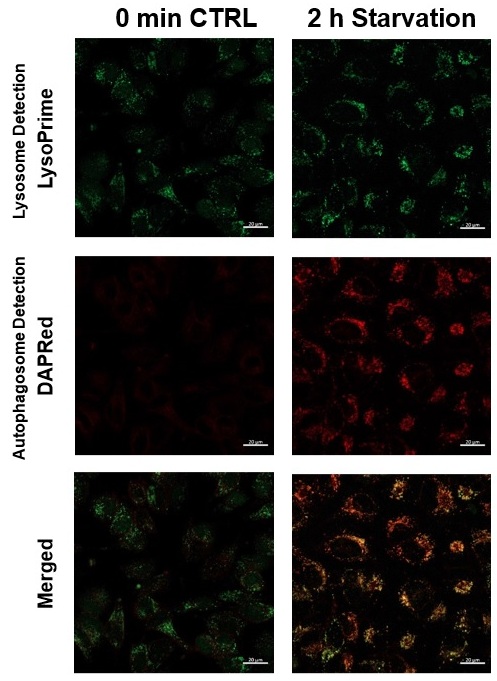
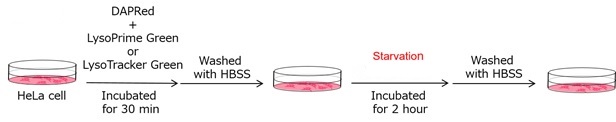
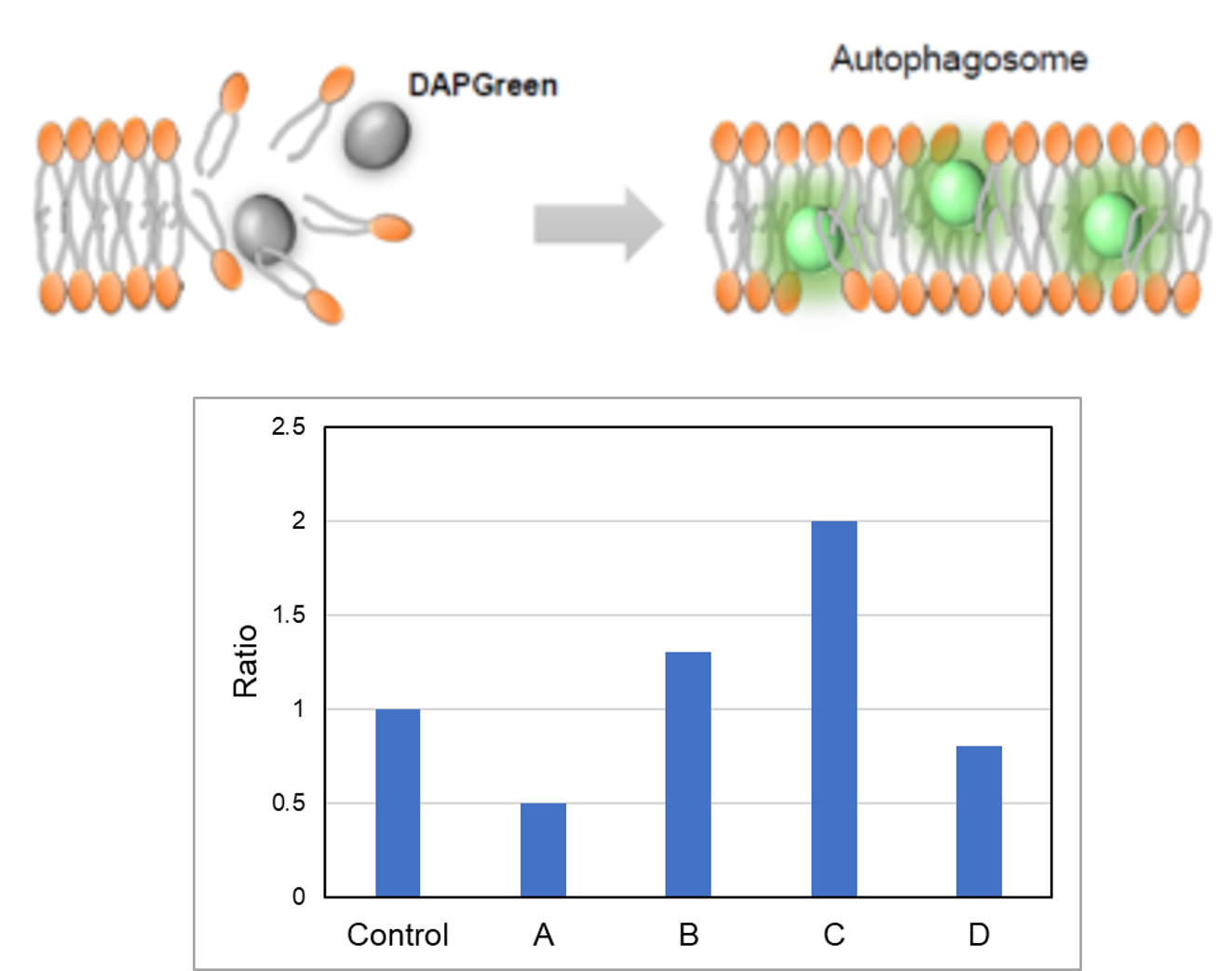
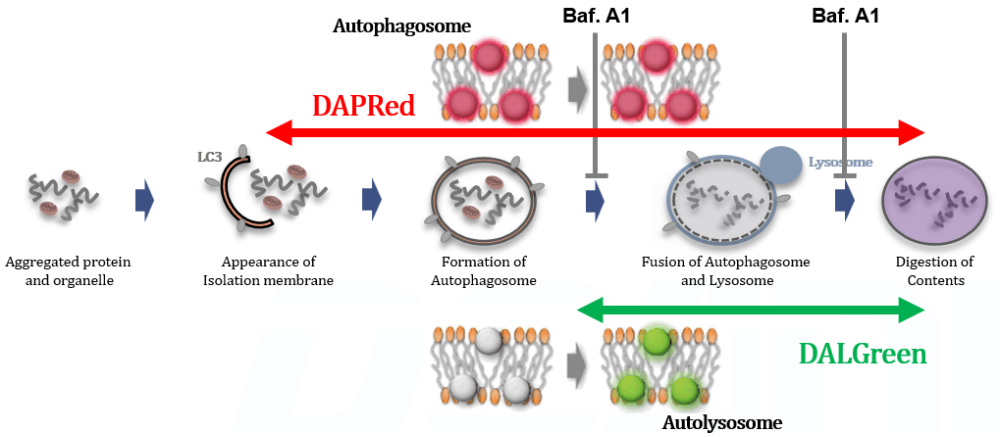
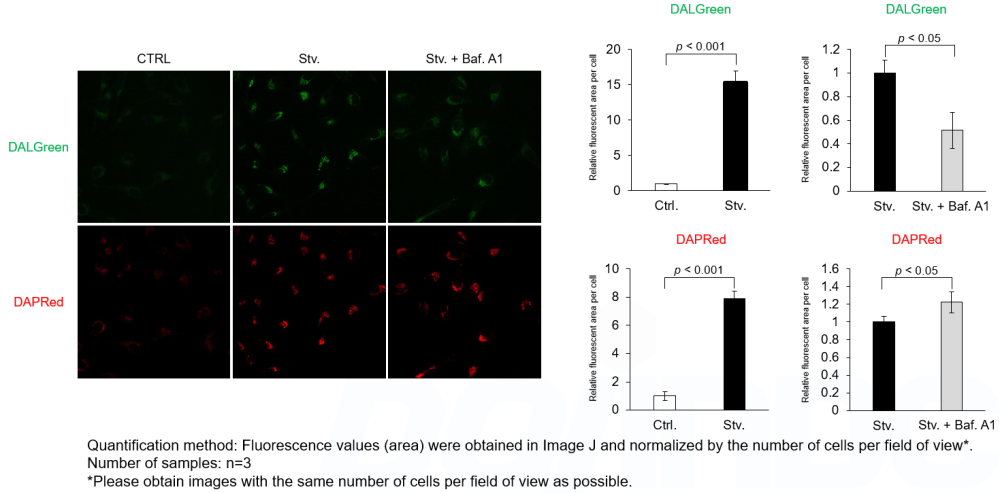
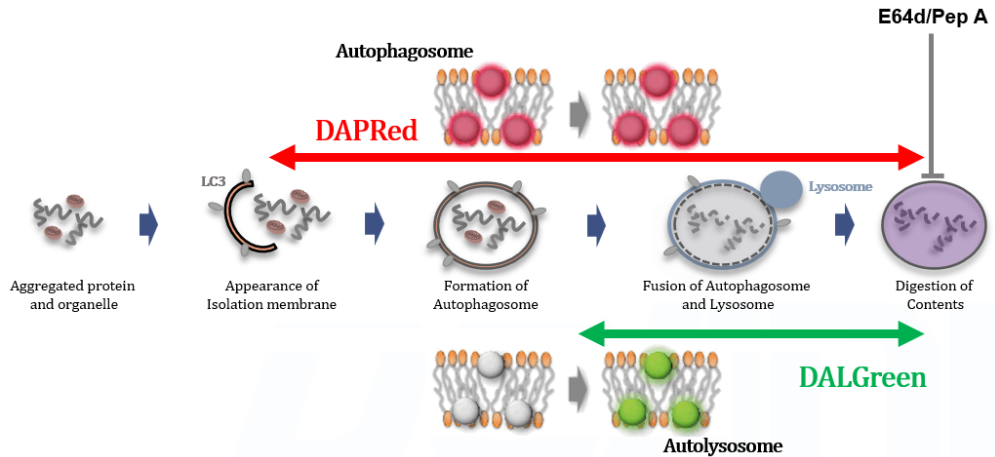
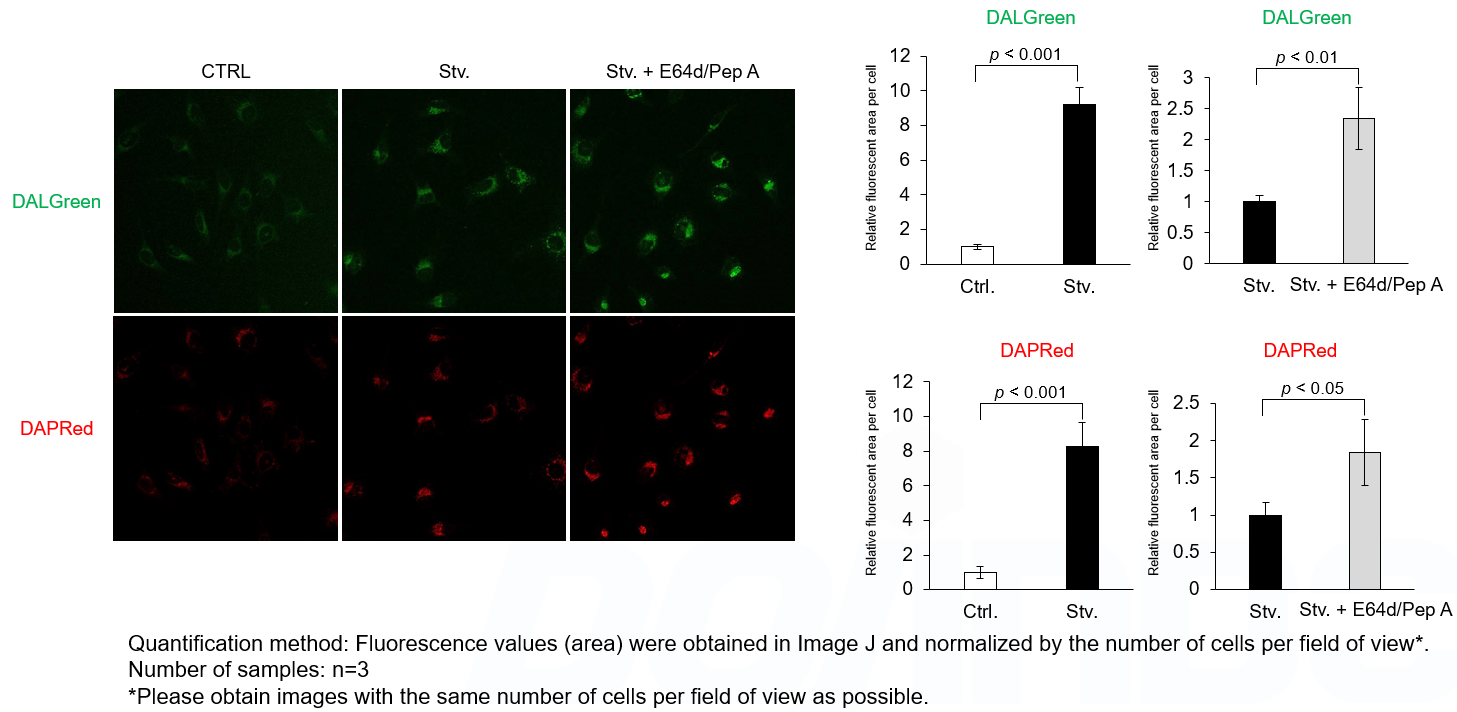
We performed fluorescence imaging under amino acid starvation in HeLa cells stained with the autophagosome detection probe DAPRed (Code: D677) and LysoPrime Green (Code: L261). The fluorescence signal of LysoPrime Green increased, and lysosomal localization was confirmed over time. These results indicate that the co-localization rate with DAPRed fluorescence is sufficient, enabling accurate autophagy analysis. |
Application Note II (click to open/close)
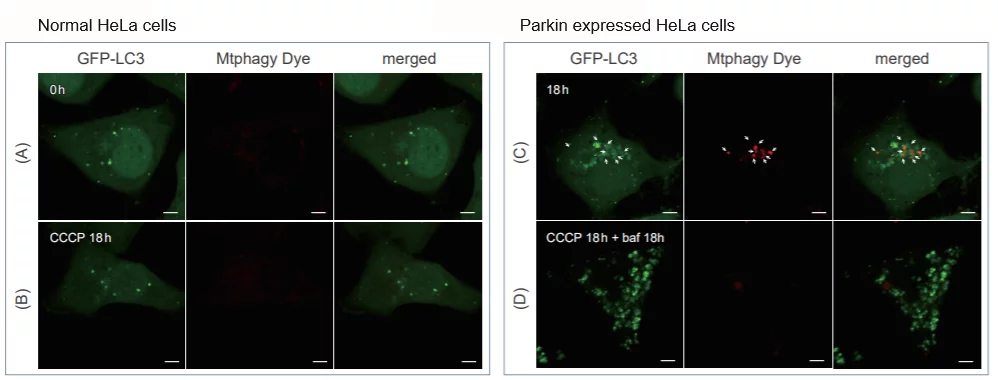
> Induction of Mitophagy in Parkin Expressed HeLa cells
|
CCCP(carbonyl cyanide m-chlorophenyl hydrazone) has been added to normal and Parkin expressed cells. The strong fluorescence was not observed in normal HeLa cells(A)(B). On the other hand, the trong fluorescence was shown in Parkin expressed cells in 18 hours after additon of CCCP(C). Some of the puncta are co-localized with the autophagy marker(GFP-LC3). In addition, suppressed fluorescence of Mtphagy dye was observed when autophagy inhibitor, bafilomycin was added to Parking expressed cells(D). Because lysosomal pH was increased by the additon of bafilomycin. |