Cellular Senescence Detection Kit - SPiDER Blue
Blue SA-β-Gal detection dye for fixed cells
- Detect SA-β-Gal levels
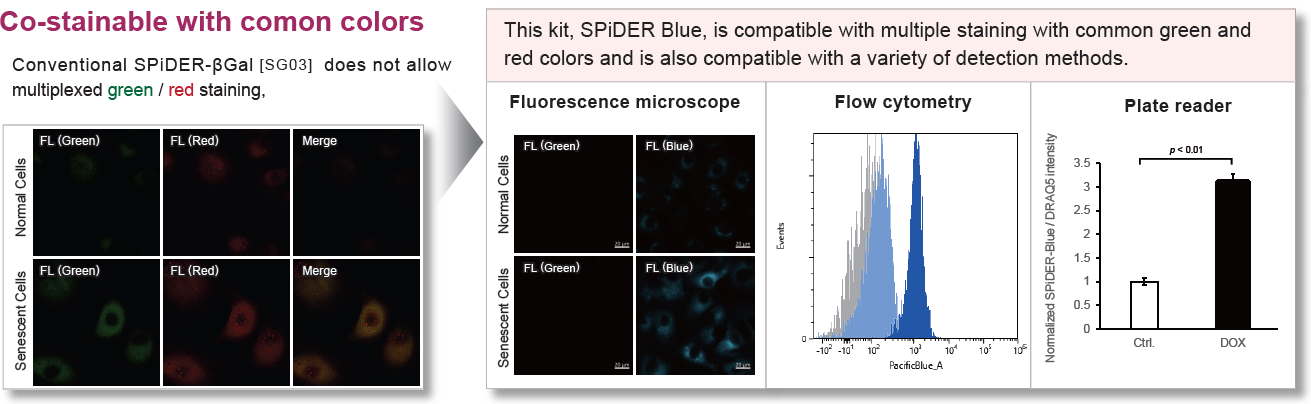
- Compatible with co-staining with immunostaining
- Enable multiple staining with common green and red dyes
-
Product codeSG07 Cellular Senescence Detection Kit - SPiDER Blue
| Unit size | Price | Item Code |
|---|---|---|
| 1 plate | Find your distributors | SG07-10 |
| 1 plate | ・SPiDER Blue ・Assay Buffer |
x 1 x 1 |
|---|
Description
This kit contains the blue fluorescent dye SPiDER Blue, which employs the detection principle of the conventional β-galactosidase detection reagent SPiDER-βGal.
SPiDER Blue detects senescence-associated β-galactosidase (SA-β-Gal, Senescence-associated β-galactosidase) and enables multiple staining with common green and red dyes, which is not possible with the conventional product SPiDER-βGal. Assay Buffer is included to suppress the background from endogenous β-galactosidase.
Cellular Senescence Analysis Products
| Product Name | Detection | Sample | Dyes / Fluorescence Properties |
|---|---|---|---|
| Cellular Senescence Detection Kit - SPiDER-βGal | Microscopy or FCM | Living / Fixed cells | SPiDER-βGal Ex: 500–540 nm / Em: 530-570 nm |
| Cellular Senescence Detection Kit - SPiDER Blue | Microscopy, FCM or Plate reader | Fixed cells | SPiDER Blue Ex: 350-450 nm / Em: 400-500 nm |
| SPiDER-βGal | Microscopy | Tissue | SPiDER-βGal Ex: 500–540 nm / Em: 530-570 nm |
| Cellular Senescence Plate Assay Kit - SPiDER-βGal | Plate reader | Living cells | SPiDER-βGal Ex: 500–540 nm / Em: 530-570 nm |
Manual
Technical info
DNA damage in normal cells may be caused by repeated cell division and oxidative stress. Cellular senescence, a state of irreversible growth arrest, can be triggered in response to DNA damage. Senescence-associated β-galactosidase (SA-β-gal), which is overexpressed in senescent cells, has been widely used as a marker of cellular senescence 1, 2).
This kit allows for detecting SA-β-gal with high sensitivity and ease of use. Because SPiDER Blue emits blue fluorescence after reacting with SA-β-gal in fixed cells, it is possible to co-stain with green or red fluorescent probes and fluorescent-labeled antibodies for immunostaining.
1) Dimri, G. P. et al., Cell Biology, 1995, 92, 9363–9367.
2) Park, A. M. et al., J. Biol. Chem., 2018, 293(41), 15815-15826.
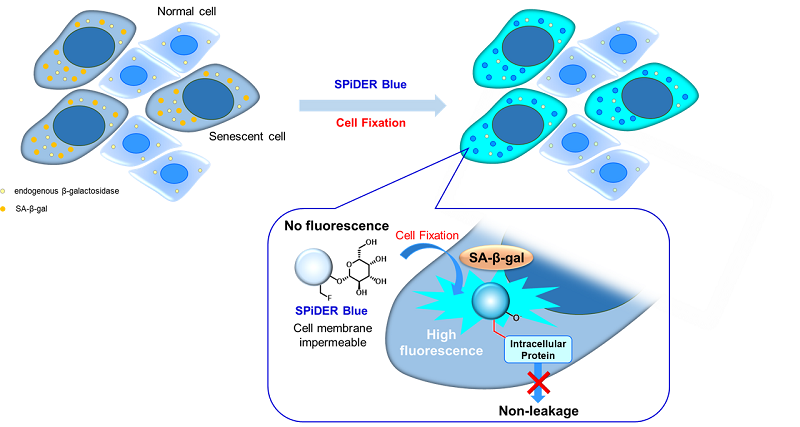
Fig. 1 Detection mechanism of senescent cells by SPiDER Blue
Fluorescence properties (SPiDER Blue after reaction with β-galactosidase)
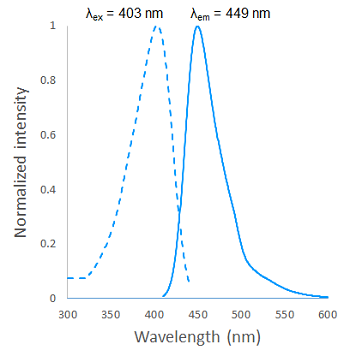
Fig. 2 Excitation and emission spectra of SPiDER Blue after reaction with β-galactosidase
Comparison of SPiDER dyes
| Product name | Feature | Living cell | Fixed cell | Detection methods | Wavelengths |
| Cellular Senescence Detection Kit SPiDER Blue |
Enables detailed analysis by multiple staining with immunostaining | - | ✓ |  |
Ex=350-450 nm Em=400-500 nm |
| Cellular Senescence Detection Kit SPiDER-βGal |
Can be used for both live cells and fixed cells, ideal for first time detection | ✓ | ✓ |  |
Ex=500-540 nm Em=530-570 nm |
| Cellular Senescence Plate Assay Kit SPiDER-βGal |
✓ | - |  |
Ex=500-540 nm Em=530-570 nm |
Experimental example: Multiple staining with oxidative stress-related markers using Doxorubicin-induced senescent cells(flow cytometry)
Using A549 cells induced to senescence by doxorubicin (DOX) and normal cells (CTRL), changes in oxidative stress-related markers in senescent cells were analyzed by flow cytometry with multiple staining. SA-βGal as a senescence marker was detected by Cellular Senescence Detection Kit - SPiDER Blue, total ROS as an oxidative stress marker was detected by ROS Assay Kit - Photo-oxidation Resistant DCFH-DA-, and γH2AX as a DNA damage marker was detected by DNA Damage Detection Kit - γH2AX-Red. As a result, total ROS and γH2AX were increased in SA-βGal-positive senescent cells, and the increase in oxidative stress-related markers associated with cellular senescence could be detected by multiple staining.
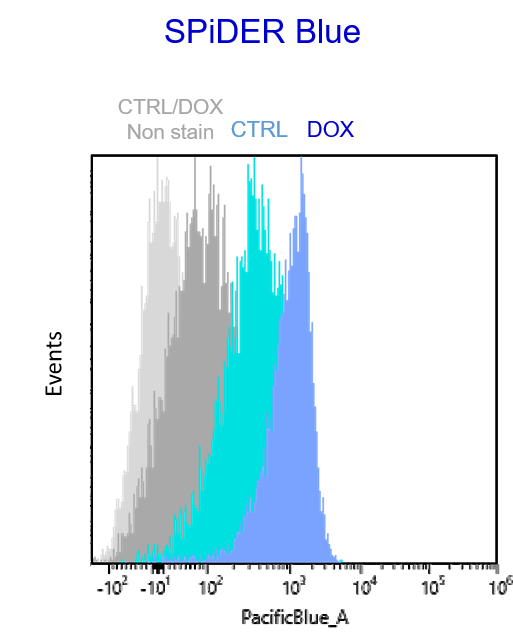
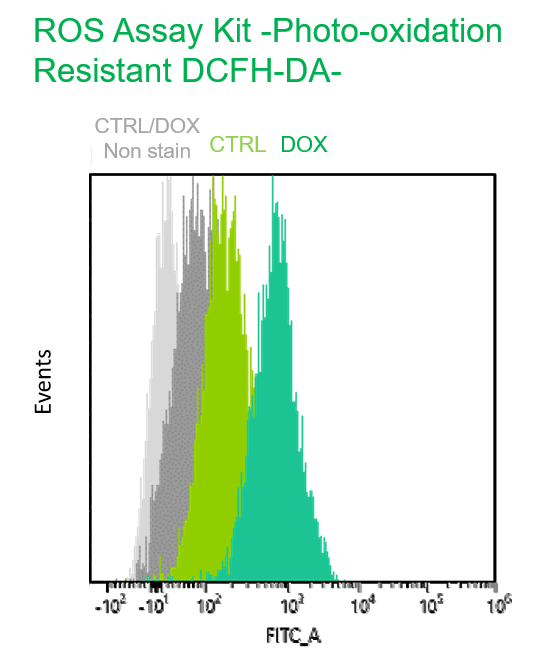
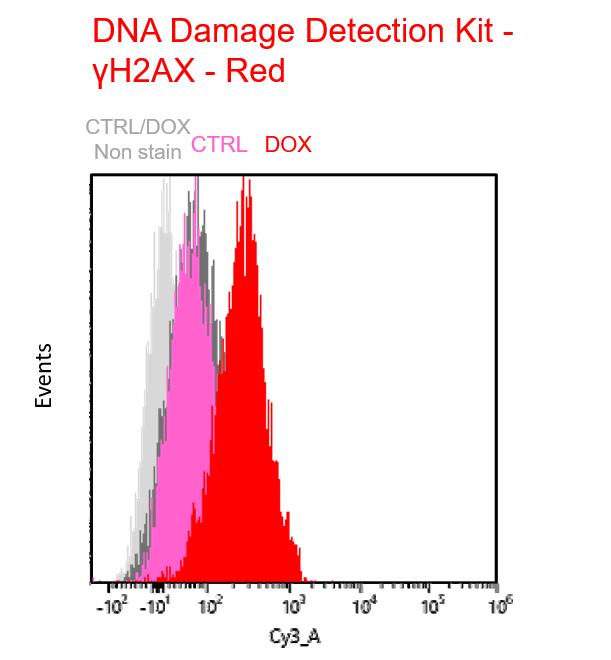
【Detection conditions】
Flow cytometry:SONY SA3800
SPiDER Blue: PacificBlue
Photo-oxidation Resistant DCFH-DA: FITC
γH2AX – Red: Cy3
<Experimental Procedure>
*Cellular senescence was induced in A549 cells by DOX (0.2 μM DOX for 3 days → normal medium for 3 days)
1. Prepare a cell suspension of A549 cells (5 x 105 cells/ml)
2. Put 1 ml of the cell suspension into a 1.5 ml tube and suspend the cells by pipetting.
3. Centrifuge at 300 x g for 5 min.
4. Remove culture medium, add 500 μl of Photo-oxidation Resistant DCFH-DA Dye Working Solution, and suspend by pipetting.
5. The suspension was incubated at 37°C for 30 minutes and centrifuged at 300 x g for 5 minutes.
6. Remove the supernatant, add 200 μl of 4% PFA/PBS solution, and incubate at 4°C for 30 min or overnight.
7. Centrifuge at 1,000 x g for 5 min.
8. Remove the supernatant, add 500 μl of 15 μmol/l SPiDER Blue solution (Assay Buffer), and suspend by pipetting.
9. The suspension was incubated at 37°C for 30 minutes and centrifuged at 1,000 x g for 5 minutes.
10. The supernatant was removed, and 100 μl of 0.1% TritonX-100 solution (PBS) was added and suspended by pipetting.
11. The suspension was incubated at room temperature for 20 minutes and centrifuged at 1,000 x g for 5 minutes.
12. Remove the supernatant, add 300 μl of Blocking Solution, and suspend by pipetting.
13. The suspension was incubated at room temperature for 20 minutes, and centrifuged at 1,000 x g for 5 minutes.
14. Remove the supernatant and add 500 μl of Anti γH2AX antibody staining solution. 15. The mixture was incubated at room temperature for 1 hour and centrifuged at 1,000 x g for 5 minutes.
16. Remove the supernatant and add 500 μl of Secondary antibody-red staining solution.
17. Incubate at room temperature for 1 hour and centrifuge at 1,000 x g for 5 minutes. 18. The supernatant was removed, and 500 μl of PBS was added and suspended by pipetting.
19. Centrifuge at 1,000 x g for 5 minutes.
20. Remove the supernatant and add 500 μl of PBS.
21. The sample was passed through a cell strainer.
22. Samples were measured on a flow cytometer (SONY: SA3800).
Experimental example: Co-staining with Lipid droplet and SA-β-Gal in fixed cells
Imaging analysis of lipid droplet accumulation in senescent cells was performed using normal A549 cells (CTRL) and cells induced senescence by Doxorubicin treatment (DOX). SA-β-Gal was detected as a senescence marker with Cellular Senescence Detection Kit - SPiDER Blue, and lipid droplets were detected with Lipi-Deep Red.
As a result, the signal of Lipi-Deep Red was increased in SA-β-Gal-positive senescent cells.
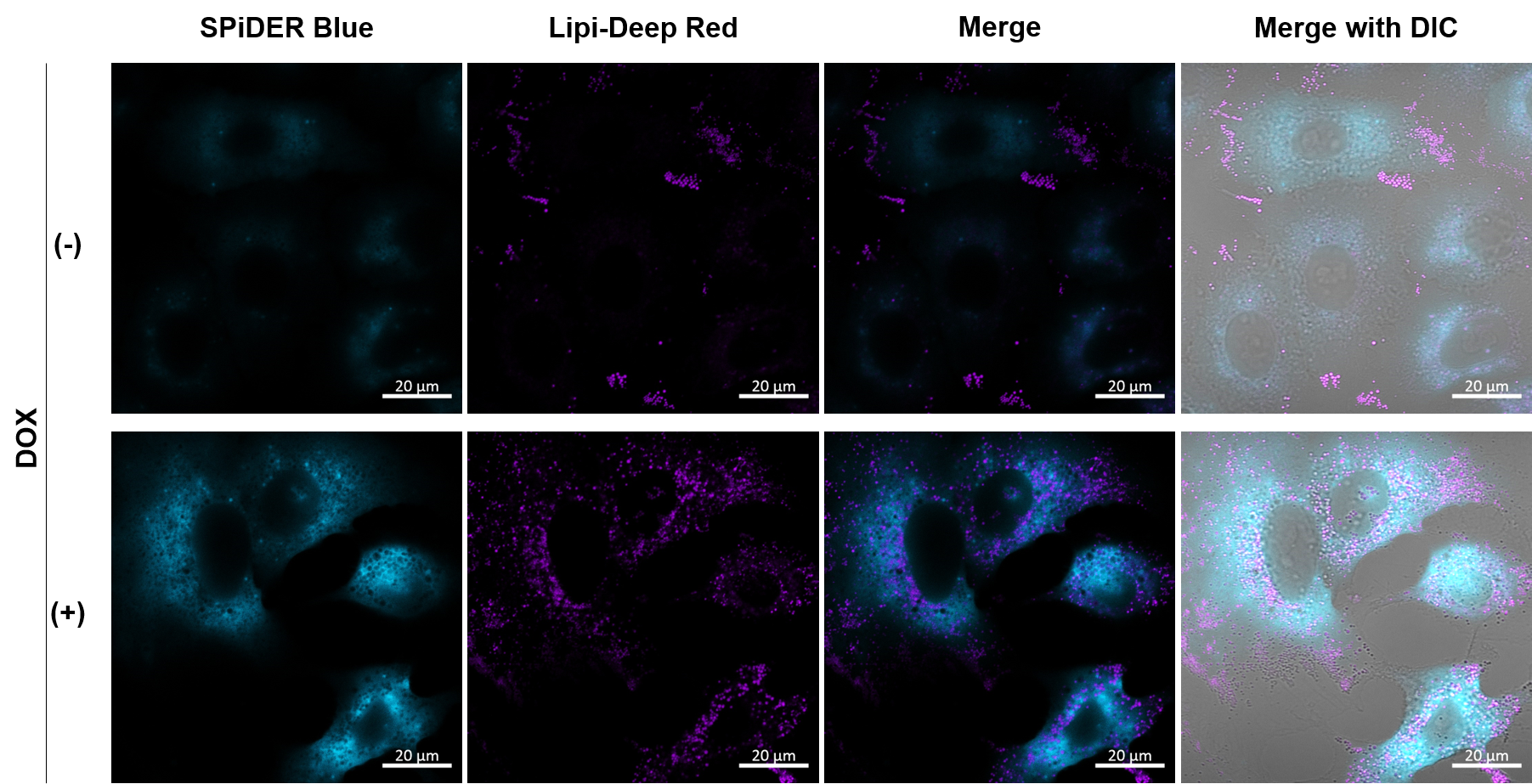
[Detection conditions]
SPiDE Blue: 405 nm (Ex), 400–550 nm (Em), 1.0%, 600V
Lipi-Deep Red: 633 nm (Ex), 650–700 nm (Em), 1.0%, 650V
*Cellular senescence was induced in A549 cells by DOX (0.2 μM DOX for 3 days → normal medium for 3 days)
1. A549 (2 x 104) cells were seeded onto µ-slide 8 well plates (ibidi) and cultured overnight in a 37°C CO2 incubator.
2. The supernatant was removed, washed once with PBS, and fixed in 4% paraformaldehyde (PFA)/PBS solution for 30 minutes at room temperature.
3. The supernatant was removed and the cells were washed once with PBS.
4. 15 µM SPiDER Blue + 0.1 µM Lipi-Deep Red prepared in Assay buffer was added and incubated at 37°C for 30 min.
5. The supernatant was removed, washed once with PBS, and 200 µl of PBS was added and observed under a confocal laser microscope (60x magnification).
Experimental example: Quantitative analysis with a fluorescent plate reader
1. A549 cells (2 x 104) were seeded onto 96 well Black Plates (Clear bottom) and cultured overnight in a 37°C CO2 incubator.
2. The supernatant was removed and 100 µl of PBS was added to wash the cells.
3. Add 100 µl of 4% PFA/PBS solution and incubate at room temperature for 30 minutes.
4. Remove the supernatant, add 100 μl of PBS, and wash the cells.
5. Add 100 μl of 15 μmol/l SPiDER Blue solution (diluted in Assay Buffer) and incubate at 37°C for 30 minutes.
6. Remove the supernatant, add 100 μl of PBS, and wash the cells.
7. Add 100 μl of 5 μmol/l DRAQ5 solution (PBS) and incubate at 37°C for 5 minutes.
8. The supernatant was removed, and cells were washed with 100 μl of PBS and measured with a fluorescent plate reader.
*To correct for cell number, nucleic acid staining was performed using DRAQ5, and fluorescence values were measured. (bottom reading)
9. Lysis Buffer*1 100 μl was added to each well and incubated at room temperature for 10 minutes.
10. Add 100 μl of Stop Solution*2 to each well.
11. The signal of SPiDER Blue was detected by a fluorescence plate reader. (top reading)
*1 Lysis Buffer: 20 mM citric acid, 40 mM phosphate buffer pH 6.0, 0.05% NP-40
*2 Stop Solution: 0.5 M sodium carbonate-sodium bicarbonate buffer pH 10.0
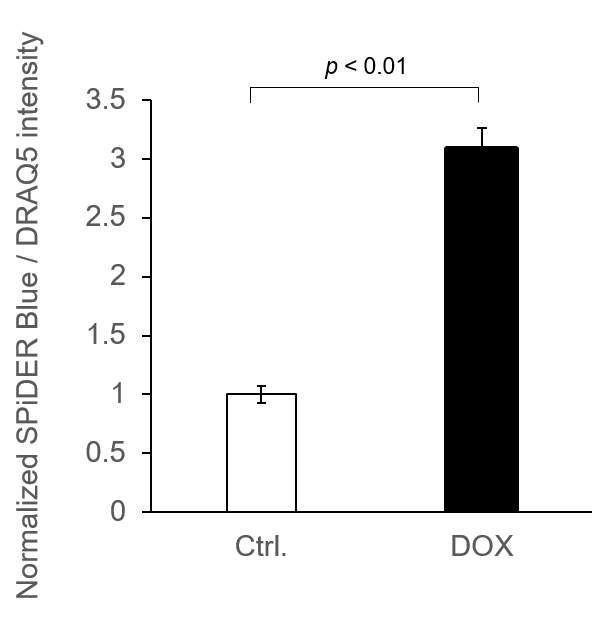
*N=3
*Normalized by DRAQ5 signals
SPiDER Blue: Ex/Em = 405/460 nm
DRAQ5: Ex/Em = 633/681 nm
Experimental example: Staining of liver adipose tissue sections from aged mice (comparison with SPiDER-βGal)
1. 8-week-old and 35-week-old mouse liver adipose tissue (frozen sections) samples were prepared on glass slides.
2. After washing once with PBS, 200 µl of 4% paraformaldehyde (PFA)/PBS solution was added and fixed at room temperature for 30 minutes.
3. The supernatant was removed and washed once with PBS.
4. Add 200 µl of 15 µM SPiDER Blue and 15 µM SPiDER-βGal prepared in Assay buffer and incubate at 37°C for 2 hours.
5. The supernatant was removed and washed once with PBS.
6. Add 1 drop of encapsulant (ProLong Glass Antifade Mountant, Thermo) and encapsulate with cover glass.
7. Observed under a confocal laser microscope (40x magnification).
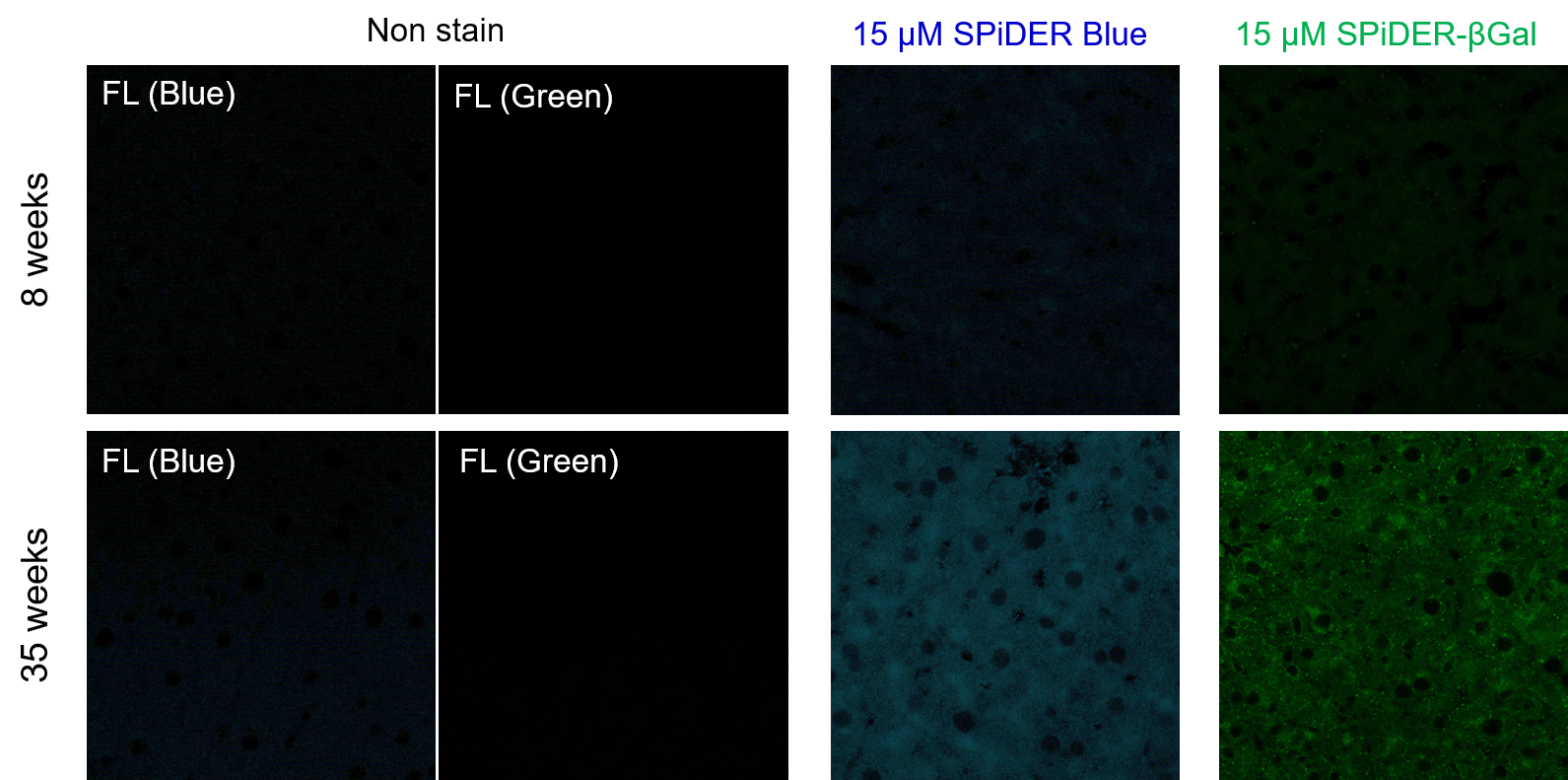
[Detection conditions]
SPiDER Blue: 405 nm (Ex), 400–500 nm (Em), 2.0%, 700V
SPiDER-βGal: 488 nm (Ex), 500–600 nm (Em), 1.0%, 600V
Experimental example: Co-staining with Mitochondrial Membrane Potential (MMP)
SA-β-Gal (SG07: SPiDER) and MMP (MT09: MT-1) were detected in human microglial cells (control) and its senescence induced cells by microscopy. Senescence induction resulted in decreased MMP and increased fluorescence of SPiDER Blue in human microglial cells.
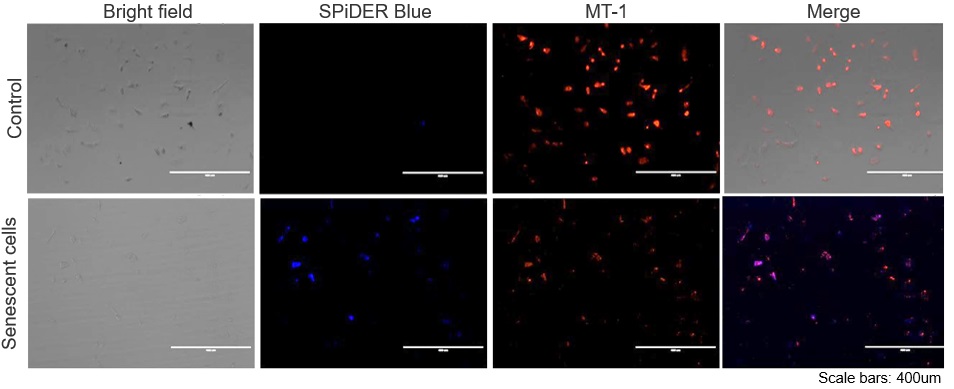
*This data was kindly provided by Dr. Supriya D. Mahajan, Department of Medicine, Jacobs School of Medicine & Biomedical Sciences.
1. Seed human microglia cells into a dish and incubate in an incubator set at 37 ℃ and equilibrated with 95% air and 5% CO2.
2. Dilute the MT-1 Dye (1:1000) in the cell culture medium.
3. Add MT-1 working solution to cells.
4. Incubate the cells for 30 minutes in an incubator set at 37 ℃ and equilibrated with 95% air and 5% CO2.
5. Discard the supernatant and wash the cells with HBSS twice.
6. Add 4% PFA/ PBS solution to the cells and incubate at room temperature for 30 minutes.
7. Discard the 4% PFA / PBS solution and wash the cells with PBS.
8. Add 15 µmol/l Spider Blue working solution and incubate at 37°C for 30 minutes.
9. Remove the working solution, and wash the cells with PBS.
10. Add Imaging Buffer solution and observe the cells under a fluorescence microscope.
Q & A
-
Q
How stable is Working solution?
-
A
Working solution cannot be stored and should be used up on the day it is prepared.
-
Q
Is it possible to store the sample after staining for later observation?
-
A
We have observed A549 cells stained with 15 μM SPiDER Blue in DOX-treated senescent cells and stored at 4°C for 3 days.
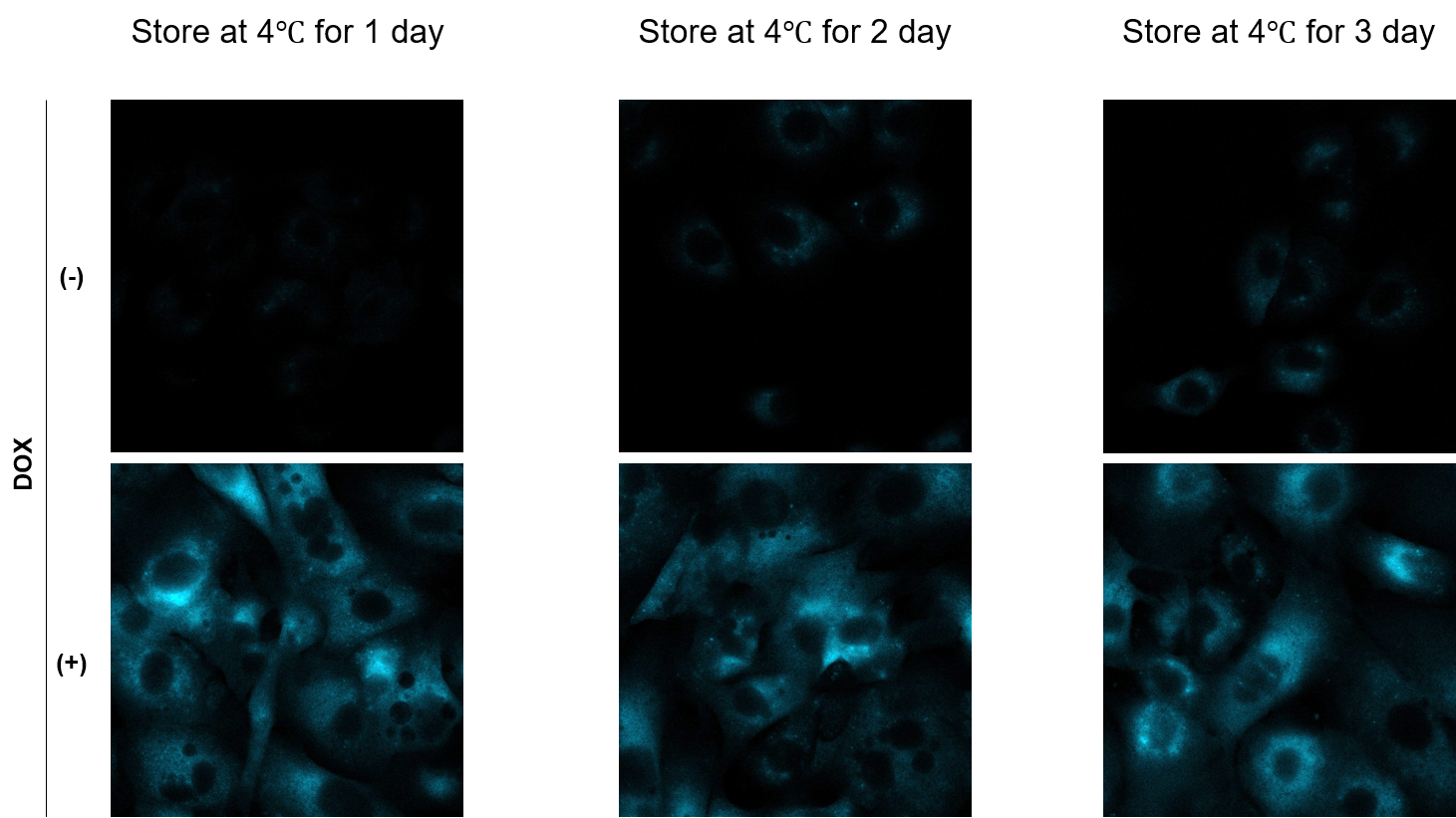
-
Q
Is it possible to observe with a fluorescence microscope?
-
A
Yes. We have experience in observation with a fluorescence microscope (KEYENCE).
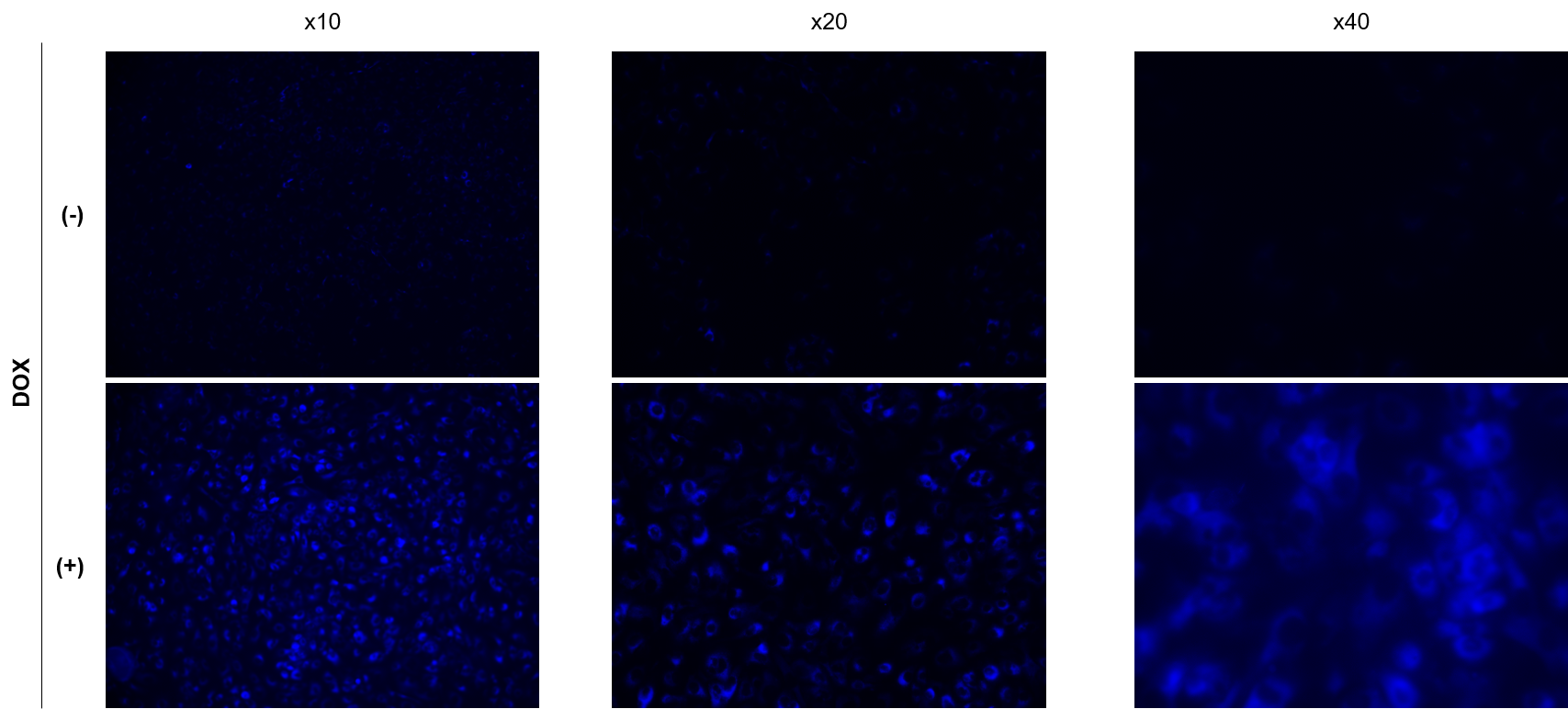
SPiDER Blue: 15 µM
Exposure time:1s (Ex = 340 - 380 nm, Em = 435 - 485 nm)
Handling and storage condition
|
Danger / harmful symbol mark |

|
|---|









