General Information
DNA damages in normal cells are caused by repeated cell division and oxidative stress. Cellular Senescence, a state of irreversible growth arrest, can be triggered in order to prevent DNA-damaged cells from growing. Senescenceassociated β-galactosidase (SA-β-gal), which is overexpressed in senescent cells, has been widely used as a marker of cellular senescence. Although X-gal staining is widely used to detect SA-β-gal, this method has the following disadvantages: 1) requirement of fixed cells due to the poor cell-permeability, 2) low quantitative capability because of the difficulty of discrimination between stained cells and unstained cells, 3) requirement of a long time for staining.
Cellular Senescence Detection Kit - SPiDER-βGal allows to detect SA-β-gal with high sensitivity and ease of use.SPiDER-βGal is a new reagent to detect β-galactosidase, and possesses high cell-permeability and intracellular retentivity. SA-β-gal in living cells can be specifically detected by using a reagent, Bafilomycin A1, to inhibit endogeneous β-galactosidase activity. In addition, SA-β-gal in fixed cells is also detectable by using McIlvaine buffer (pH 6.0). Since SPiDER-βGal emits strong and stable fluorescence after the reaction with SA-β-gal, it can be applied to quantitative analysis by flow cytometry.
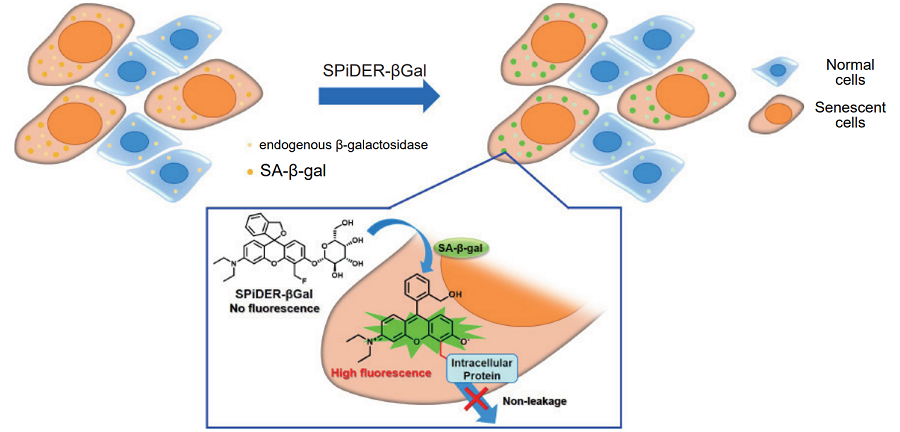
Fig. 1 Detection mechanism of senescent cells by SPiDER-βGal
< Recommended filter >
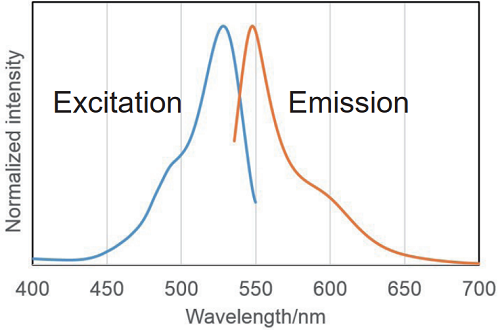
Ex:500 ~ 540 nm
Em:530 ~ 570 nm
There are internal detection examples obtained using confocal microscopy and flow cytometry at an excitation wavelength of 488 nm.
Fig. 2 Excitation and emission spectra of SPiDER-βGal after reaction with β-galactosidase
Kit Contents
10 assays (35 mm dish)
|
SPiDER-βGal |
×1 |
|
Bafilomycin A1 |
×1 |
- The Bafilomycin A1 in the tube may be barely visible due to the small amount. Please handle it carefully.
Storage Condition
Store at 0-5℃
Required Equipment and Materials
- Dimethyl sulfoxide (DMSO)
- Culture medium or HBSS
- Micropipettes
Preparation of Solutions
Preparation of SPiDER-βGal DMSO stock solution
Add 14 μl of DMSO to a tube of SPiDER-βGal and dissolve it with pipetting. Store the SPiDER-βGal DMSO stock solution at -20℃.
Preparation of Bafilomycin A1 DMSO stock solution
Add 24 μl of DMSO to a tube of Bafilomycin A1 and dissolve it with pipetting. Store the Bafilomycin A1 DMSO stock solution at -20℃.
- Insoluble substances such as lipofuscin accumulate in cells according to the acceleration of cellular senescence. Because lipofuscin has autofluorescence, it may results in higher fluorescence background. In order to achieve accurate SA-β-gal activity assay in senescent cells, we recommend preparing samples without SPiDER-βGal staining and compare the fluorescence intensities of both cells with or without SPiDER-βGal staining. Please refer to our web page or contact technical service for details.
General Protocol
- Assay for living cells -
Preparation of Bafilomycin A1 working solution
Dilute the Bafilomycin A1 DMSO stock solution 1,000 times with culture medium or HBSS.
Preparation of SPiDER-βGal working solution
Mix the SPiDER-βGal DMSO stock solution and Bafilomycin A1 DMSO stock solution. Dilute the mixture 1,000 times with culture medium or HBSS.
- For example, in order to prepare 1 ml of the SPiDER-βGal working solution, mix 1 μl of SPiDER-βGal DMSO stock solution and 1 μl of Bafilomycin A1 DMSO stock solution. Dilute the mixture with 1 ml of culture medium or HBSS.
- Prepare cells on 35 mm dish for assay and culture the dish at 37℃ overnight in a 5% CO2 incubator.
- Discard the culture medium and wash the cells with 2 ml of culture medium or HBSS once.
- Add 1 ml of Bafilomycin A1 working solution and incubate at 37℃ for 1 hour in a 5% CO2 incubator.
- Add 1 ml of SPiDER-βGal working solution and incubate at 37℃ for 30 minutes in a 5% CO2 incubator.
- After removing the supernatant, wash the cells with 2 ml of culture medium or HBSS twice.
- Observe the cells under a fluorescence microscope or analyze by a flow cytometer.
- Assay for fixed cells -
Preparation of SPiDER-βGal working solution
Dilute the SPiDER-βGal DMSO stock solution 2,000 times with McIlvaine buffer (pH 6.0).
- Preparation of McIlvaine buffer (pH 6.0):
Mix 0.1 mol/l citric acid solution (3.7 ml) and 0.2 mol/l sodium phosphate solution (6.3 ml). Confirm the pH is 6.0. If the pH is not 6.0, adjust the pH by adding either citric acid solution or sodium phosphate solution. Dilute this buffer 5 times with ultrapure water.
- Prepare cells on 35 mm dish for assay and culture the dish at 37℃ overnight in a 5% CO2 incubator.
- After removing the culture medium, wash the cells with 2 ml of HBSS once. Add 2 ml of 4%paraformaldehyde (PFA) / PBS solution to the cells and incubate at room temperature for 3 minutes.
- Remove the supernatant, and wash the cells with 2 ml of HBSS three times.
- Add 2 ml of SPiDER-βGal working solution and incubate at 37℃ for 30 minutes.
- We recommend not to use a 5% CO2 incubator for fixed cell experiments.If incubation is done in a 5% CO2 incubator, the pH of the buffer may become acidic. Acidic pH results in higher background fromthe endogenous β-galactosidase activity and it would be difficult to distinguish between normal cells and senescent cells.
- After removing the supernatant, wash the cells with 2 ml of HBSS twice.
- Observe the cells under a fluorescence microscope or analyze by a flow cytometer.
- Examples of a flow cytometry analysis are available in the product web page.
Usage Examples
Fluorescence imaging of SA-β-gal
-
- WI-38 cells (5×104 cells/dish, MEM, 10%fetal bovine serum, 1%penicillin-streptmycin) of passage number 0 and 12 were seeded respectively in a μ-dish 35 mm (ibidi) and cultured overnight in a 5% CO2 incubator.
- The cells were washed with 2 ml of HBSS once.
- Bafilomycin A1 working solution (1 ml) was added to the culture dish, and the cells were incubated for 1 hour in a 5% CO2 incubator.
- SPiDER-βGal working solution (1 ml) and 1 mg/ml Hoechst 33342 (1 μl) were mixed. Then the mixture solution was added to the culture dish, and the cells were incubated for 30 minutes in a 5% CO2 incubator.
- After the supernatant was removed, the cells were washed with 2 ml of HBSS twice.
- HBSS (2 ml) were added and the cells were observed by confocal fluorescence microscopy (Excitation: 488 nm, Emission: 500-600 nm).
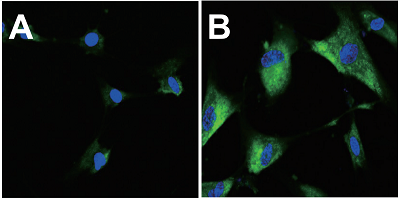
A. Passage 0, B. Passage 12
(green: SPiDER-βGal, blue: Hoechst 33342)
Fig. 3 Fluorescence imaging of SA-β-gal in WI-38 cells
Quantitative analysis of SA-β-gal positive cells by flow cytometry
-
- WI-38 cells (1×105 cells/dish, MEM, 10%fetal bovine serum, 1% penicillin-streptmycin) of passage number 1 and 12 were seeded respectively in a μ-dish 35 mm (ibidi) and cultured overnight in a 5% CO2 incubator.
- The cells were washed with 2 ml of HBSS once.
- Bafilomycin A1 working solution (1 ml) was added to the culture dish, and the cells were incubated for 1 hour in a 5% CO2 incubator.
- SPiDER-βGal working solution (1 ml) was added to the culture dish, and the cells were incubated at for 30 minutes in a 5% CO2 incubator.
- After the supernatant was removed, the cells were washed with 2 ml of HBSS twice.
- The cells were harvested by trypsin and resuspended in MEM (10%fetal bovine serum, 1%penicillin-streptmycin).
- The cells were observed by a flow cytometer (Excitation: 488 nm, Emission: 515-545 nm).
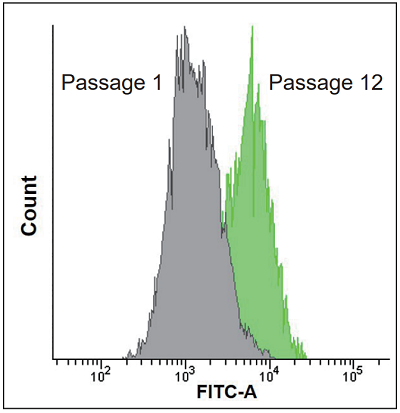
Fig. 4 Quantification of SA-β-gal positive WI-38 cells
Reference
- T. Doura, M. Kamiya, F. Obata, Y. Yamaguchi, T. Y. Hiyama, T. Matsuda, A. Fukamizu, M. Noda, M. Miura and Y. Urano, Angew. Chem. Int. Ed., 2016, 55, 9620.
Frequently Asked Questions / Reference
SG03: Cellular Senescence Detection Kit - SPiDER-βGal
Revised Nov., 16, 2023


 Hidden sections will not be printed.
Hidden sections will not be printed.