pHLys Red - Lysosomal Acidic pH Detection

Lysosomal pH Detection Reagent Red
- High Sensitivity
- High Specificity
- Able to Overnight Staining
-
Product codeL265 pHLys Red - Lysosomal Acidic pH Detection
| Unit size | Price | FUJIFILM Wako Products code |
|---|---|---|
| 1 tube | ¥28,000 | 349-10011 |
| 3 tubes | ¥48,000 | 345-10013 |
35 mm dish ×10, μ-Slide 8 well ×10, 96-well Plate ×2 for each tube.
Description
The lysosome is an organelle in which a biomembrane forms an acid vacuole. Lysosomes contain various degrading enzymes and contribute to maintaining intracellular homeostasis by acting as a waste disposal system. Recent findings reveal that lysosomal dysfunction is related to some neurodegenerative disorders. Consequently, the investigation of lysosomal function is attracting considerable interest in the scientific community.
Live imaging using small molecule fluorescent probes has been widely used for lysosomal live cell analysis, but the low specificity and retention ability due to pH change have been cited as issues. The pHLys Red is a small molecule fluorescent probe with high lysosomal specificity and sensitivity to pH changes, enabling a more accurate analysis of lysosomal pH in live cells. It is also applicable to experiments that require long-term imaging due to its high retention ability.
Lysosomal Analysis Products
| Product Name | Lysosomal pH Detection Dyes and Fluorescence Properties |
Lysosomal Quantity Detection Dyes and Fluorescence Properties |
|---|---|---|
| Lysosomal Acidic pH Detection Kit | pHLys Red Ex: 561 nm / Em: 560-650 nm |
LysoPrime Green Ex: 488 nm / Em: 500-600 nm |
| Lysosomal Acidic pH Detection Kit - Green/Deep Red | pHLys Green Ex: 488 nm / Em: 490-550 nm |
LysoPrime Deep Red Ex: 633 nm / Em: 640-700 nm |
| pHLys Red - Lysosomal Acidic pH Detection | pHLys Red Ex: 561 nm / Em: 560-650 nm |
|
| LysoPrime Deep Red - High Specificity and pH Resistance | LysoPrime Deep Red Ex: 633 nm / Em: 640-700 nm |
|
| LysoPrime Green- High Specificity and pH Resistance | LysoPrime Green Ex: 488 nm / Em: 500-600 nm |
Manual
Technical info
Competitor's dye for lysosomal pH detection has issues in accurate lysosomal localization and sensitivity. Fluorescence imaging of HeLa cells treated with low concentrations of the lysosomal acidity inhibitor Bafilomycin A1 (Baf. A1) and stained with pHLys Red or competitor's dye was compared. The results showed that pHLys Red was more sensitive to changes in lysosomal pH than the competitor's dye, indicating that pHLys Red is more sensitive to changes in lysosomal pH than the competitor's dye.
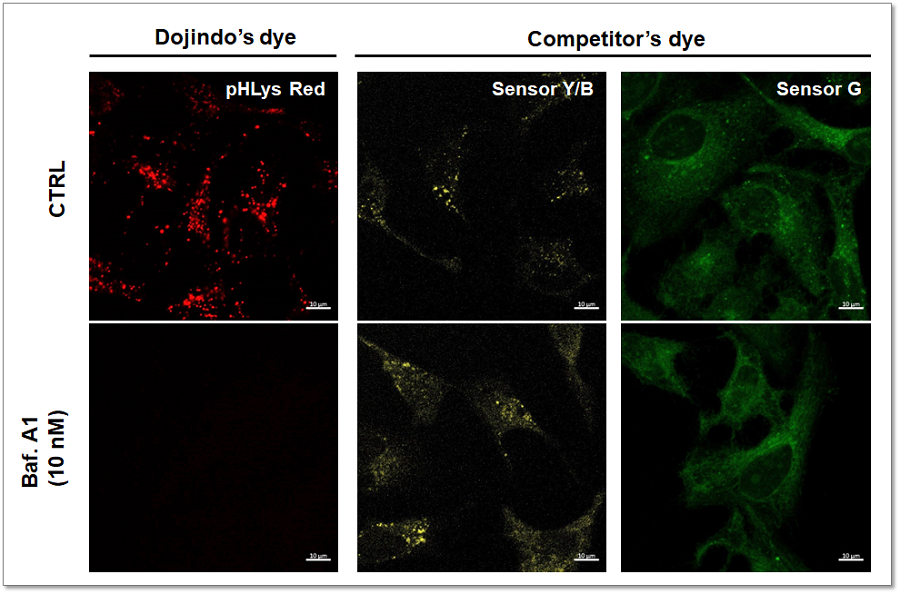
<Experimental Conditions>
pHLys Red:Ex = 561 nm, Em = 560 - 620 nm
Sensor Y/B:Ex = 561 nm, Em = 560 - 620 nm
Sensor G:Ex = 488 nm, Em = 490 - 550 nm
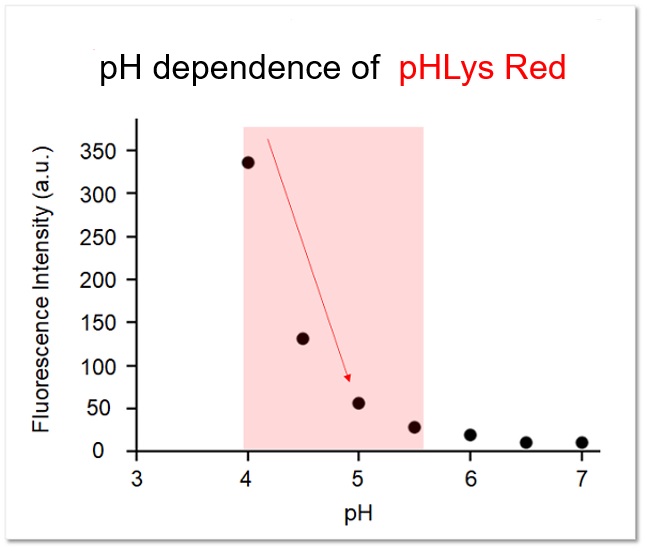
The fluorescence intensity of pHLys Red at each pH was confirmed in vitro, and it was confirmed that the fluorescence intensity changed sensitively within the range of lysosomal pH (pH 4.0-5.5).
High Specificity for Lysosomes
The lysosomal localization of competitor's dye and pHLys Red was compared using HeLa cells expressing the lysosomal marker protein LAMP1-GFP. The competitor's dye stained HeLa cells showed higher background due to the leakage from lysosomes, whereas pHLys Red showed lower background (Merged image). The pHLys Red was found to be more accurately localized to lysosomes compared to the competitor's dye.
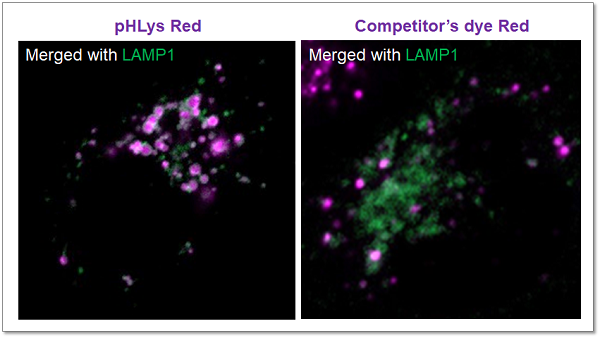
<Experimental Conditions>
Green: Ex= 488 nm, Em= 500-570 nm
Red : Ex= 561 nm, Em= 560-620 nm
High Retention in Lysosome
The lysosomal retention ability of pHLys Red and competitor's dye was compared using cells stained with each dye. The fluorescence of the competitor's dye decreased 24 hours after staining, whereas the fluorescence of pHLys Red was maintained and retention was high.
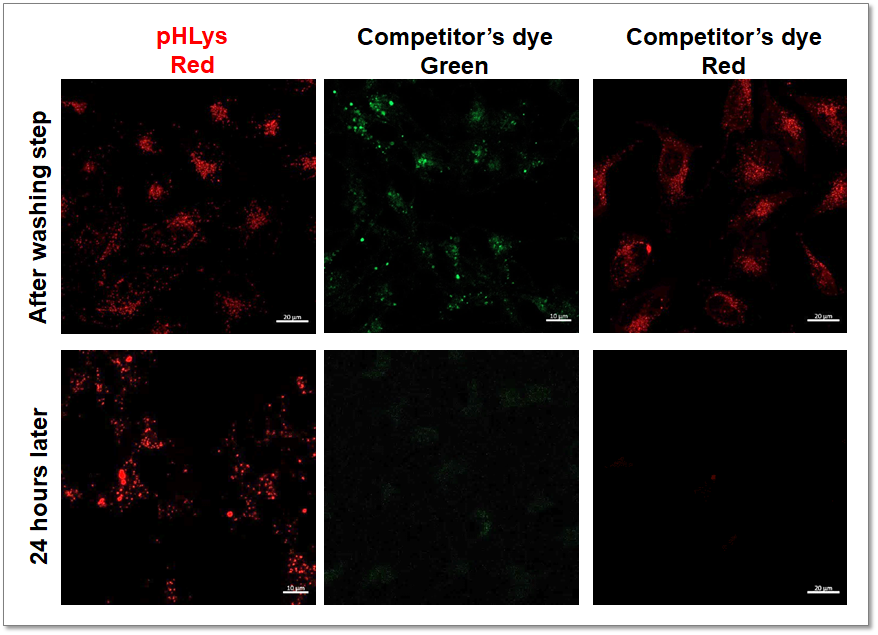
<Experimental Conditions>
Green: Ex= 488 nm, Em= 490-550 nm
Red: Ex= 561 nm, Em= 560-620 nm
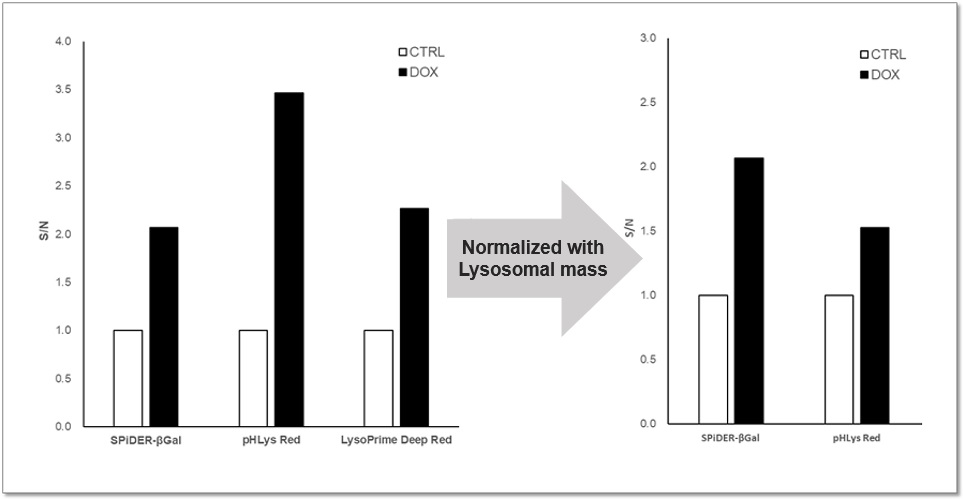
Application Data: Lysosomal Mass and pH Exchange in Senescence-induced Cells
We analyzed senescence-associated acidic β-galactosidase (SA-βGal) activity and lysosomal mass and pH in A549 cells treated with Doxorubicin (DOX) to induce senescence. The SA-βGal activity was detected by SG03 Cellular Senescence Detection Kit - SPiDER-βGal, the lysosomal mass and pH were detected separately with L264 LysoPrime Deep Red and pHLys Red. Fluorescence imaging showed that the increase in lysosomal mass and pH acidification were observed in senescence-induced cells, and the normalized fluorescence intensity of lysosomal mass and pH by plate reader measurement showed the same result.

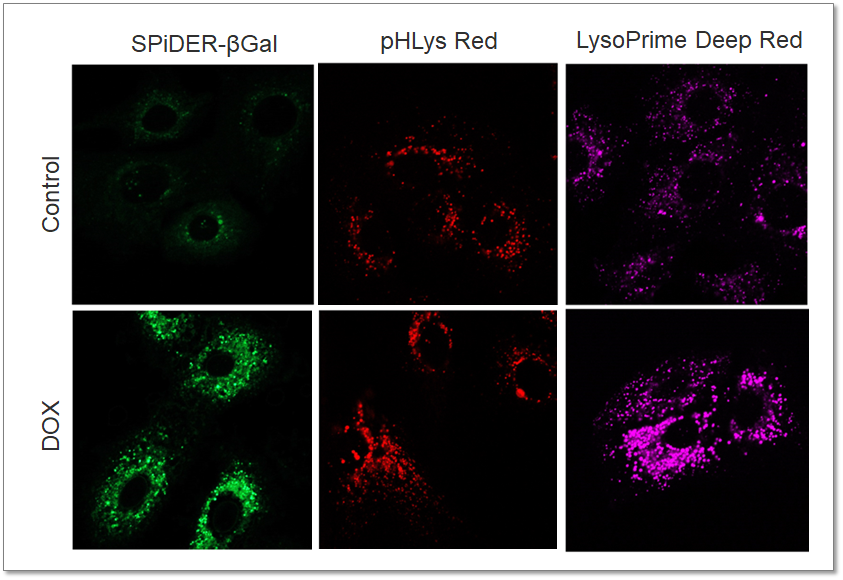
<Experimental Conditions>
SA-βGal(Green):
Ex = 488 nm, Em = 490 – 550 nm
Lysosomal pH (Red):
Ex = 561 nm, Em = 560 – 620 nm
Lysosomal mass (Deep Red):
Ex = 633 nm, Em = 640 – 700 nm
<Experimental Conditions>
SA-βGal: Ex = 525 – 535 nm, Em = 550 – 570 nm
Lysosomal pH: Ex = 555 – 565 nm, Em = 590 – 610 nm
Lysosomal mass: Ex = 645 – 655 nm, Em = 690 – 710 nm
References
| No. | Cell Type | Inducer | Instrument | Reference (Link) |
|---|---|---|---|---|
| 1) | NK-92 cells | Ammonia (NH₄Cl), Bafilomycin A1 | Microscope | J. Domagala, T.M. Grzywa, I. Baranowska, M. Justyniarska, R. Tannir, A. Graczyk-Jarzynka, A. Kusowska, M. Lecka, M. Poreba, K. Fidyt, K. Marhelava, Z. Pilch, L.K. Picard, T. Wegierski, K. Jastrzebski, M. Krawczyk, M. Klopotowska, M. Granica, D. Urlaub, S. Hajduk, A. Neeser, S. Moros, P. Kozlowski, M. Bobrowicz, M. Miaczynska, L. Ma, C. Watzl, M. Winiarska, "Ammonia Suppresses the Antitumor Activity of Natural Killer Cells and T Cells by Decreasing Mature Perforin", Cancer Res, 2025, doi:10.1158/0008-5472.CAN-24-0749 |
| 2) | GBM cells | Nigericin, Monensin | Flow cytmeter | Y. Jing, M. Kobayashi, M. I. Shoulkamy, M. Zhou, H. T. Vu, H. Arakawa, H. Sabit, S. Iwabuchi, C. Q. Vu, A. Kasahara, M. Ueno, Y. Tadokoro, K. Kurayoshi, X. Chen, Y. Yan, S. Arai, S. Hashimoto, T. Soga, T. Todo, M. Nakada, A. Hirao, "Lysine-arginine imbalance overcomes therapeutic tolerance governed by the transcription factor E3-lysosome axis in glioblastoma", Nat. Commun., 2025, doi:10.1038/s41467-025-56946-z |
| 3) | Bone marrow-derived macrophages | LPS, rhFGF21, Bafilomycin A1 | Microscope | J. Zhu, Z. Jin, J. Wang, Z. Wu, T. Xu, G. Tong, E. Shen, J. Fan, C. Jiang, J. Wang, X. Li, W. Cong, L. Lin, "FGF21 ameliorates septic liver injury by restraining proinflammatory macrophages activation through the autophagy/HIF-1α axis", J Adv Res, 2025, doi:10.1016/j.jare.2024.04.004 |
| 4) | HeLa cell | Bafilomycin A1, Monensin | Microscope | F. Kraus, Y. He, S. Swarup, K.A. Overmyer, Y. Jiang, J. Brenner, C. Capitanio, A. Bieber, A. Jen, N.M. Nightingale, B.J. Anderson, C. Lee, J.A. Paulo, I.R. Smith, J.M. Plitzko, S.P. Gygi, B.A. Schulman, F. Wilfling, J.J. Coon, J.W. Harper, "Global cellular proteo-lipidomic profiling of diverse lysosomal storage disease mutants using nMOST", Sci Adv, 2025, doi:10.1126/sciadv.adu5787 |
| 5) | U2OS cell | Activation of TRPML1, Bafilomycin A1 | Microscope | C. Lee, M.J.G. Eldridge, M.A. Gonzalez-Lozano, T. Bresnahan, Z. Niday, D. Del Camino, T. Fu, J.A. Paulo, M.M. Moran, S. Helaine, J.W. Harper, "DMXL1 promotes recruitment of V1-ATPase to lysosomes upon TRPML1 activation", Nat Struct Mol Biol, 2025, doi:10.1038/s41594-025-01581-x |
| 6) | HeLa-FcγRII cell | Ammonia (NH₄Cl), Bafilomycin A1, infection | Microscope | T. Nakamura, T. Shimizu, N. Nishinakama, R. Takahashi, K. Arasaki, A. Uda, K. Watanabe, M. Watarai, "A novel method of Francisella infection of epithelial cells using HeLa cells expressing fc gamma receptor", BMC Infect Dis, 2024, doi:10.1186/s12879-024-10083-y |
| 7) | Human CB CD34+ cell | Different culture conditions | Flow cytmeter | K. Ishitsuka, H. Nishinakama, T. Kimura, A. Sugiyama-Finnis, "Purging myeloma cell contaminants and simultaneous expansion of peripheral blood-mobilized stem cells", Exp Hematol, 2024, doi:10.1016/j.exphem.2023.07.769 |
| 8) | Adipocytes differentiated from MSCs | Genetic deletion or pharmacological inhibition of Scd1 | Microscope | H. Mori, S.K. Peterson, R.C. Simmermon, K.A. Overmyer, A. Nishii, E. Paulsson, Z. Li, A. Jen, R.M. Uranga, J.N. Maung, W.T. Yacawych, K.T. Lewis, R.L. Schill, T. Hetrick, R. Seino, K. Inoki, J.J. Coon, O.A. MacDougald, "Scd1 and monounsaturated lipids are required for autophagy and survival of adipocytes", Mol Metab, 2024, doi:10.1016/j.molmet.2024.101916 |
Q & A
-
Q
Can I use other solutions to prepare working solution instead of medium?
-
A
Yes, you can use HBSS or PBS to prepare the working solution instead of the medium.
-
Q
When should I add the stimulation? Before the pHLys Red staining or after?
-
A
We recommend adding the stimulation after the pHLys Red staining. The pHLys Red accumulates in normal lysosomes in a pH-dependent manner. However, the pHLys Red cannot accumulate in lysosomes if the pH has changed to near neutral due to the drug stimulation. Therefore, drug stimulation after staining is recommended.
For stimulation with Bafilomycin A1, you can add the Bafilomycin A1 into the pHLys Red working solution or add the Bafilomycin A1 after the pHLYs Red staining step.
-
Q
I want to evaluate the lysosomal mass and pH at the same time, can I use LysoPrime to co-staining?
-
A
Yes, you can. However, please notice that you should stain the cells with LysoPrime first, and then stain with pHLys Red.
-
Q
Which instruments is applicable for pHLys Red?
-
A
pHLys Red is applicable for Fluorescent microscopes, Confocal microscopes, and plate readers.
・Confocal microscope
Ex = 561 nm, Em = 560-620 nm・Fluorescent microscope
TxRed Filter
Ex = 540 - 580 nm, Em: 590 - 670 nm・Plate Reader
Ex = 555 – 565 nm, Em = 590 – 610 nm
-
Q
What is the recommended final concentration of pHLys Red?
-
A
We recommend using a 1,000-fold dilution of pHLys Red, but if you want to optimize the staining conditions, please refer to the following range.
Please consider using a dilution of 250-500x.
-
Q
Can cells be fixed after staining with pHLys Red?
-
A
Fixation is not recommended.















