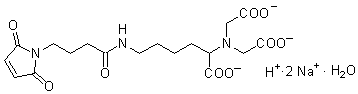
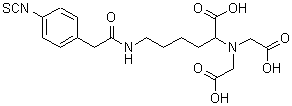
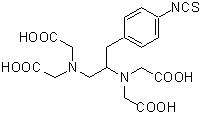FeBABE
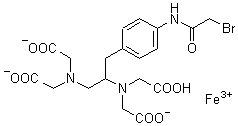
Reagent for Biopolymer Function Analysis
-
Product codeF279 FeBABE
-
CAS No.186136-50-5
-
Chemical name1-(p-Bromoacetamidobenzyl)ethylenediamine-N,N,N',N'-tetraacetic acid, iron(III)
-
MWC19H21BrFeN3O9=571.13
| Unit size | Price | Item Code |
|---|---|---|
| 1 mg | Find your distributors | F279-10 |
Description
Product Description of BABEs
Bromoacetamidobenzyl-EDTA (BABE) is a chelate labeling reagent that conjugates with sulfhydryl groups. The iron chelate of BABE (FeBABE) is a unique tool for determining the three-dimensional structure of proteins and the binding structures of protein-protein or protein-DNA complexes. BABE adds EDTA moieties to proteins through their sulfhydryl groups. Once attached to a protein, FeBABE cuts a nearby peptide or DNA chain. The cleavage site is within 12 angstroms of the FeBABE binding site. Iron (II)-chelate cleaves a peptide or DNA chain in the presence of hydrogen peroxide. The cleavage reaction completes quickly: 10 seconds to 20 minutes of incubation is sufficient. The size of the cleaved fragment is analyzed with gel electrophoresis such as SDS-PAGE.
References
1) L. H. DeRiemer, C. F. Meares, D. A. Goodwin and C. I. Diamanti, "BLEDTA II: Synthesis of a New Tumer-Visualizing Derivative of Co(III)-bleomycin", J. Labelled Compd. Radiopharm., 1981, 18, 1517.
2) T. M. Rana and C. F. Meares, "Specific Cleavage of a Protein by an Attached Iron Chelate", J. Am. Chem. Soc., 1990, 112, 2457.
3) T. M. Rana and C. F. Meares, "Transfer of Oxigen from an artificial protease to peptide carbon during proteolysis", Proc. Natl. Acad. Sci. USA, 1991, 88, 10578.
4) D. P. Greiner, R. Miyake, J. K. Moran, A. D. Jones, T. Negishi, A. Ishihama and C. F. Meares, "Synthesis of the Protein Cutting Reagent Iron (S)-1-(p-Bromoacetamidobenzyl)ethylebediaminetetraacetate and Conjuation to cysteine Sie Cahins", Bioconjugate Chem., 1997, 8, 44.
5) E. Platis, M. R. Ermacora and R. O. Fox, "Oxidative Polypeptide Cleavage Mediated by EDTA-Fe Covalently Linked to Cysteine Residue", Biochemistry, 1993, 32, 12761.
6) S. L. Traviglia, S. A. Datwyler, D. Yan, A. Ishihama and C. F. Meares, "Targeted Protein Footprinting: Where Different Transcription Factors bind to RNA Polymerase", Biochemistry, 1999, 38, 4259.
7) J. B. Ghaim, D. P. Greiner, C. F. Meares and R. B. Gennis, "Proximity Mapping the suface of Membrane Protein Using an Artificial Protease: Demonstration That the Quinone-Binding Domain of Subunit I Is near the N-Terminal Region of Subunit II of Cytochrome bd", Biochemistry, 1995, 34, 11311.
8) R. Miyake, K. Murakami, J. T. Owens, D. P. Greiner, O. N. Ozoline, A. Ishihama and C. F. Meares, "Dimeric Association of Escherichia coli RNA Polymerase alfa subunits, studied by Cleavage of Single-Cysteine alfa Sununits Conjugated to Iron-(S)-1-(p-(Bromoacetamido)benzyl)ethylenediaminetetraacetate", Biochemistry, 1998, 37, 1344.
9) J. T. Owens, R. Miyake, K. Murakami, A. J. Chmura, N. Fujita, A. Ishihama and C. F. Meares, "Mapping the sigma70 subunits contact sites on Escherichia coli RNA polymerase with a sigma70-conjugated chemical protease", Proc. Natl. Acad. Sci. USA, 1998, 95, 6021.
10) J. A. Bown, J. T. Owens, C. F. Meares, N. Fujita, A. Ishihama, S. J. Busby and S. D. Minchin, "Organization of open complexes at Escherichia coli promoters. Location of promoter DNA sites close to region 2.5 of the sigma70 subunit of RNA polymerase", J. Biol. Chem., 1999, 274, 2263.
11) F. Colland, N. Fujita, D. Kotlarz, J. A. Bown, C. F. Meares, A. Ishihama and A. Kolb, "Positioning of sigma(S), the stationary phase sigma factor, in Escherichia coli RNA polymerase-promoter open complexes", EMBO J., 1999, 18, 4049.
12) G. M. Heilek, R. Marusak, C. F. Meares and H. F. Noller, "Directed hydroxyl radical probing of 16S rRNA using Fe(II) tethered to ribosomal protein S4", Proc. Natl. Acad. Sci. USA, 1995, 92, 1113.
13) G. M. Heilek and H. F. Noller, "Site-directed hydroxyl radical probing of the rRNA neighborhood of ribosomal protein S5", Science, 1996, 272, 1659.
14) K. R. Lieberman and H. F. Noller, "Ribosomal protein L15 as a probe of 50 S ribosomal subunit structure.", J. Mol. Biol., 1998, 284, 1367.
Handling and storage condition
| Appearance: | Yellowish brown powder |
|---|---|
| Purity (HPLC): | ≧ 95.0 % |
| Solubility in water: | To pass test (clear, yellowish brown) |
| -20°C, 3.Protect from moisture |