AcidSensor Labeling Kit – Endocytic Internalization Assay
- Intracellular Fluorescent Probes
- Intracellular Fluorescent Probes
- Fluorescent dye
- Fluorescence microscope
- Labeling kit
- NH2
pH Sensor Labeling Kit
- Label with pH-sensitive fluorescent dye
- All-in-one Kit
- Detailed Labeling Manual
-
Product codeA558 AcidSensor Labeling Kit – Endocytic Internalization Assay
| Unit size | Price | Item Code |
|---|---|---|
| 3 samples | Find your distributors | A558-10 |
| Amount of Sample | 50-200 µg |
|---|---|
| Labeling Group | -NH2 |
| Instrument | Microscope/ FCM |
| Fluorescence | [Ex:645, Em:666] |
|
・Proteins with a molecular weight above 50,000 can be labeled. ・High recovery rate using a filtration tube. |
|
| 3 samples | ・NH2-Reactive AcidSensor ・WS Buffer ・Reaction Buffer ・Filtration Tube |
3 tubes 4 ml×1 500 μl×1 3 tubes |
|---|
Description
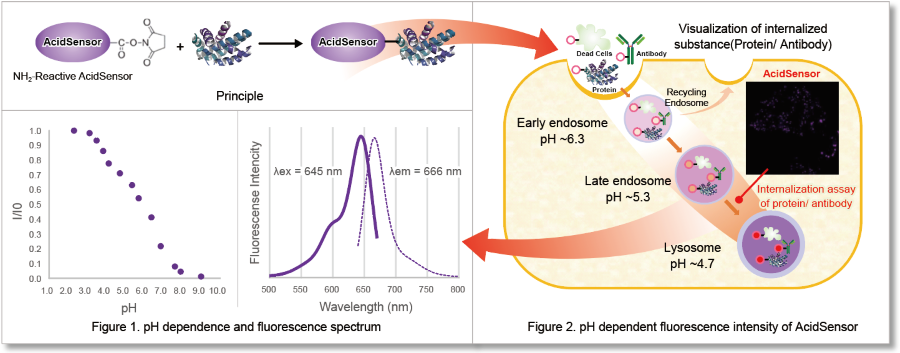
This kit is an all-in-one kit that allows visualization of the endocytosis uptake of a target substance. The NH2-Reactive AcidSensor (fluorescent probe) included in the kit has an intramolecular active ester group that forms a stable covalent bond when mixed with an amino group-containing target substance (protein). The AcidSensor label can be excited at 633 nm, allowing for multiple staining with green or red fluorescence (Figure 1). The AcidSensor label shows little fluorescence in neutral conditions and fluoresces when acidified in the cells where it is taken up by endocytosis (Figure 2).*Notice:
・Unlike the endocytosis detection dye: ECGreen (product code: E296), this kit stains target substances that enter the cell.
・This kit can label samples with molecular weights of more than 50,000 and with reactive amino groups.
Manual
Technical info
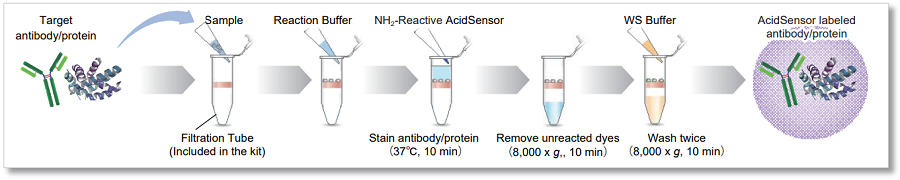
This kit includes a filtration tube necessary to remove the unreacted dye, and allows you to perform everything from labeling to purification operations. *
In addition, even first-time users can easily label AcidSensor by conducting experiments according to the instruction manual.
* Protein/Antibody is not included.
* Please prepare DMSO and a proper medium.
Compare with other Kits
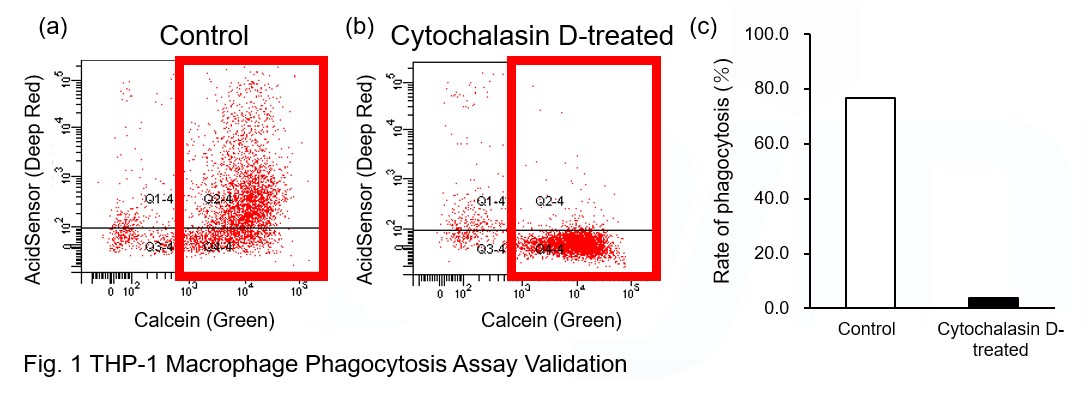
Experimental Example: Phagocytosis assay of labeled apoptotic cells in THP-1 cells
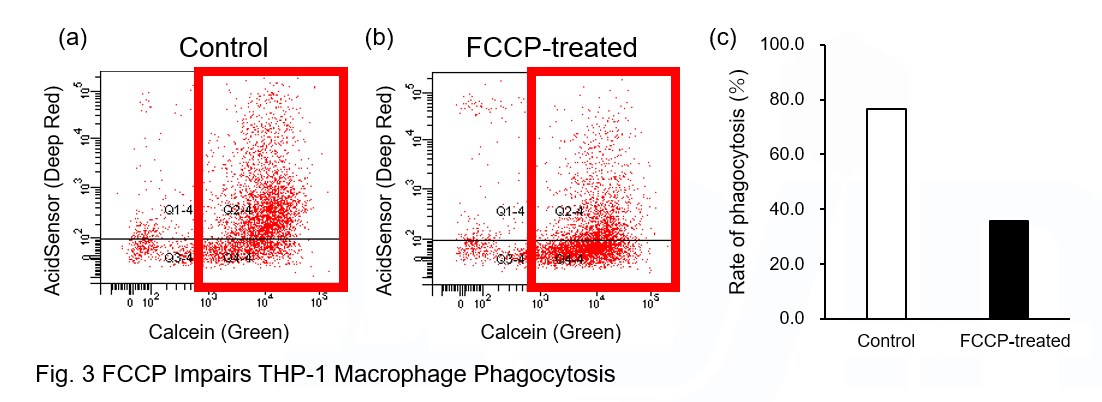
AcidSensor-labeled substances are taken up by cells and their fluorescence increases when they reach acidic organelles such as lysosomes. Taking advantage of this property, we evaluate the phagocytic activity of apoptotic cells by co-culturing AcidSensor-labeled apoptotic cells with Calcein-labeled THP-1 macrophages. As a result, Calcein (Green) / AcidSensor (Deep red) double-positive cells, indicating THP-1 macrophages phagocytosing apoptotic cells, were observed by flow cytometry (Fig. 1a). Furthermore, when the phagocytosis of THP-1 macrophages was inhibited by Cytochalasin D, the percentage of double-positive cells decreased (Fig. 1b and 1c), confirming that the assay system can accurately evaluate phagocytosis.
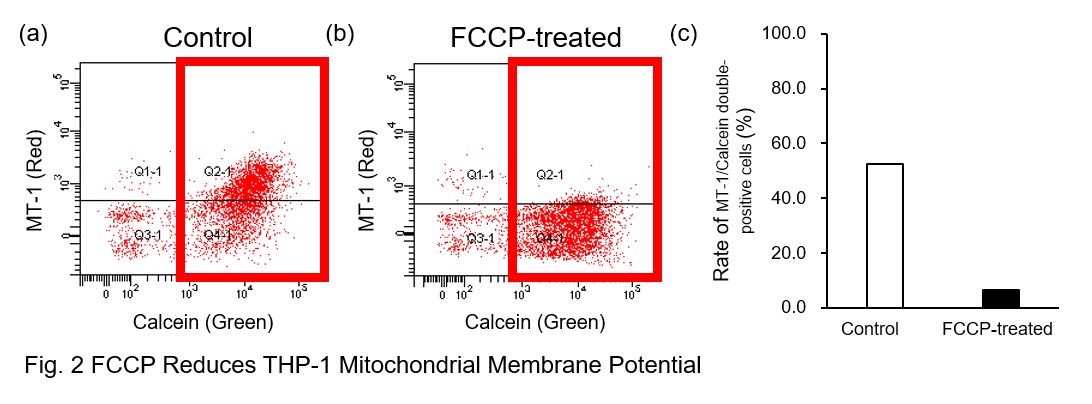
A recent report reveals that inhibition of mitochondrial function induces a switch to glycolysis and reduces phagocytosis in cultured microglia, resident macrophages in the central nervous system*. To replicate this result, phagocytosis assays were performed using mitochondria-inhibited THP-1 macrophages. The results show that FCCP, a potent uncoupler of oxidative phosphorylation in mitochondria, decreases mitochondrial membrane potential (MT-1, Red) of THP-1 macrophages (Fig. 2) and reduces phagocytosis (Fig. 3).
*Lauren H. Fairley, et al., PNAS (2023)
Products in Use
① AcidSensor Labeling Kit – Endocytic Internalization Assay [code: A558]
② -Cellstain- Calcein-AM solution [code: C396]
③ MT-1 MitoMP Detection Kit [code: MT13]
[Experimental Procedure]
Preparation of AcidSensor-labeled apoptotic cells (day before assay)
1. Add 10 μl of DMSO to NH2-Reactive AcidSensor and dissolve. 5 μl NH2-Reactive AcidSensor solution was added to 5 ml HBSS to make the Working solution (1000-fold dilution).
2. Wash Jurkat cells twice with HBSS.
3. Jurkat cells (5×107 cells) were transferred to the tube and the supernatant was removed after centrifugation.
4. Add the Working solution to the tube with Jurkat cells (5×107 cells) and suspend.
5. Incubate at 37°C for 30 minutes for labeling with AcidSensor.
6. After labeling, the cells were washed twice with HBSS.
7. AcidSensor-labeled Jurkat cells were suspended in RPMI medium with 10% FBS added with 0.5 μM staurosporine.
8. Apoptosis was induced by o/n culture in an incubator (37°C, 5% CO2) for 18 hours.
9. After induction of apoptosis, the cells were washed with a culture medium and used for the phagocytosis assay.
Phagocytosis assay using THP-1 macrophages
1. To differentiate THP-1 cells into macrophages, THP-1 cells were seeded into 6 well plates at 1x106 cells/well and incubated with 100 nM PMA for 3 days in an incubator.
2. After washing THP-1 macrophages twice with HBSS, Calcein-AM (Code: C396, 0.5 μg/ml) and MT-1 (Code: MT13, 1000-fold dilution) included HBSS solution was added to the wells and incubated in the incubator for 30 minutes.
3. Wash twice with a culture medium.
4. The cells were incubated with 10 μM Cytochalasin D for 1 hour or 5 μM FCCP for 30 minutes in the incubator.
5. After twice washing with culture medium, AcidSensor-labeled apoptotic cells (3x106 cells/well) were added to the well with or without Cytochalasin D or FCCP. The cells were incubated in the incubator for 4 hours.
6. Wash twice with HBSS.
7. Add 500 μl Imaging buffer solution (Code: MT13) and collect THP-1 macrophages from plates with a cell scraper.
8. Cell suspensions were analyzed by flow cytometry.
Co-staining for Observation with Endosomes
Cellular uptake of labeled IgG over time
AcidSensor-labeled mouse IgG stained with this kit and Dojindo's endocytosis detection dye (Code: E296, ECGreen -Endocytosis Detection)were added to HeLa cells. Observed the cells 10, 20, and 180 minutes after the staining, the results showed that the AcidSensor (Deep Red) and endosomal membranes(Green) were localized at the same place, indicating that mouse IgG was uptake by the cells via the endocytosis pathway.
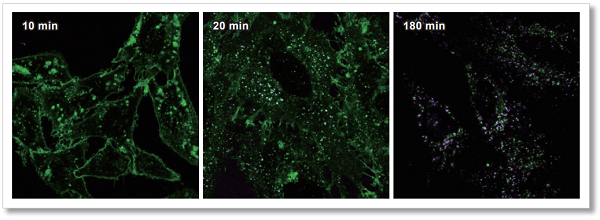
<Observation Conditions>
Green:ECGreen (Ex = 405 nm, Em = 500-550 nm)
Deep Red:AcidSensor (Ex = 633 nm, Em = 650-700 nm)
Application Data AcidSensor Labeled BSA and Mouse IgG Uptake by HeLa Cells
Labeled BSA or Mouse IgG was added to HeLa cells using this kit, and uptake by HeLa cells was observed after 2 hours. As a result, AcidSensor fluorescent was detected in HeLa cells, confirming that each labeled object was taken up by the endocytosis pathway.
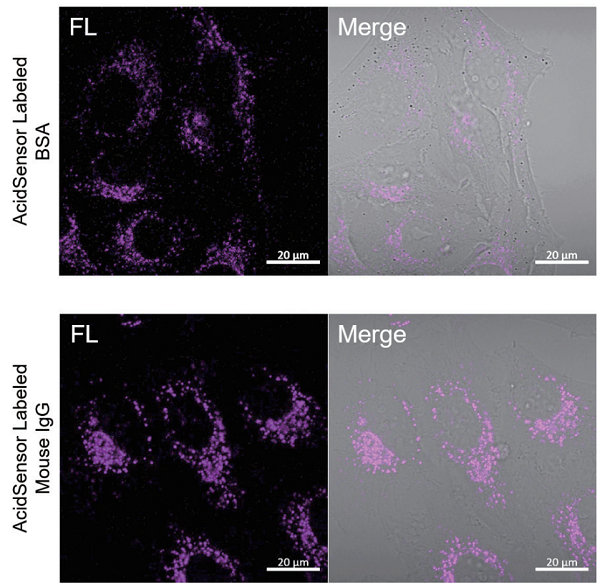
<Observation Conditions>
Instrument: Confocal Microscope
Ex = 633 nm, Em = 650-700 nm
Q & A
-
Q
Do coexisting substances in the sample solution affect the labeling reaction?
-
A
Depends on the coexisting substances.
After confirming what substances are contained in the solution, purify the sample accordingly.
Coexisting compounds with amino groups and a molecular weight above 10,000 will decrease the labeling efficiency, and cannot be removed. Even for compounds without amino groups, high molecular weight impurities can cause clogging of the filter, which may interfere with labeling and purification. Please purify the sample before using it.
-
Q
Is there any cytotoxicity in the WS buffer?
-
A
WS buffer contains a detergent in a quantity that shows almost no cytotoxicity. If you are concerned about the effect on cells, collect the labeled substance using a buffer of your choice.
-
Q
Is it possible to take up or observe a labeled substance in serum-containing medium?
-
A
Serum components do not affect AcidSensor performance, but serum proteins may inhibit the uptake of the endocytosis pathway. Please consider whether to include serum or not depending on your experiment.
-
Q
How should I calculate the labeling rate?
-
A
Dilute the labeled sample 5-fold with WS buffer, measure the absorbance at 500 nm and 280 nm, and calculate the labeling rate using the following formula.

A500: Absorbance at 500 nm
A280: Absorbance at 280 nm
εprotein: The proteins' coefficient of molar absorbance at 280nm
The A500 coefficient of molar absorbance of NH2-Reactive AcidSensor is 48400
Using the A280 and A 500 of NH2-Reactive AcidSensor, the ratio of 280nm/500nm is 0.16
For IgG samples, the εprotein is 216,000
-
Q
How many NH2-Reactive AcidSensors are labeled per protein molecule?
-
A
For Mouse IgG, we have confirmed that an average of three AcidSensors are labeled per molecule.
Handling and storage condition
| Store in a cool (0–5°C), dark, and dry place. |