Previous Science Note
|
Lysosomal Dysfunction Directly Contributes to Disease Progression Recent studies have shown that lysosomal dysfunction plays an active role in disease mechanisms. In Alzheimer’s disease, impaired lysosomal acidification prevents the clearance of amyloid-beta, leading to autophagic vacuole buildup and toxic plaque formation. In cancer, dysregulated lysosomal iron triggers lipid peroxidation and ferroptosis, offering a path to eliminate treatment-resistant cells. These findings position lysosomes as key players in both pathological progression and therapeutic response. |
||||||||||||||||||||||
|
Summary: Lysosomal dysfunction in Alzheimer’s disease impairs acidification, disrupting the breakdown of amyloid-beta (Aβ) and related proteins. As a result, autophagic vacuoles build up and form flower-like structures, leading to cell death and plaque formation. Highlighted technique: This study used a neuron-specific transgenic fluorescent protein (mRFP-eGFP-LC3) to monitor lysosomal acidification. Because eGFP loses its signal in acidic environments while mRFP remains stable, reduced acidification could be detected by comparing the two signals using confocal and electron microscopy. Related technique Accurate lysosomal detection / Autophagic Flux Assay Kit |
||||||||||||||||||||||
|
Activation of lysosomal iron triggers ferroptosis in cancer (Nature, 2025) Summary: When lysosomes lose control of iron activity, they can trigger damaging reactions that lead to ferroptosis. This hidden malfunction offers a new way to target treatment-resistant cancer cells, positioning lysosomes as a key focus in drug development. Highlighted technique: This study visualized lysosomes using LAMP1 immunostaining and Lysosome specific probes, allowing detection of structural and acidification changes. In addition, lipid peroxidation and iron accumulation were monitored together with lysosomes to evaluate their involvement in ferroptosis. Related technique Intracellular Iron Detection, Lipid Peroxide Detection (used in this article) |
||||||||||||||||||||||
All Related Techniques (click to open/close)
|
||||||||||||||||||||||
Application Note (click to open/close)
|
||||||||||||||||||||||
|
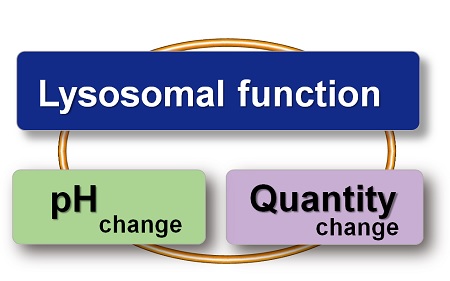
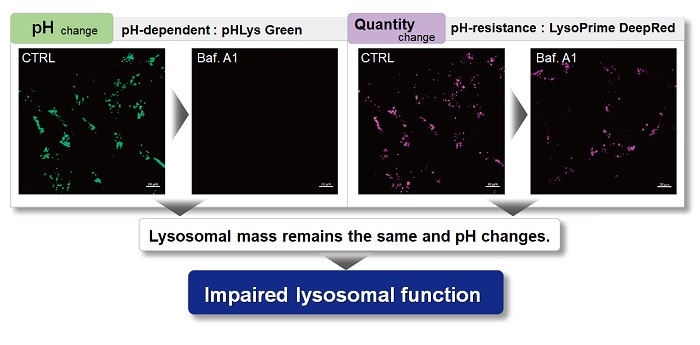
With existing reagents, it was difficult to determine whether lysosomal mass or their function (pH) fluctuated because the discussion was based on changes in the fluorescence brightness of a single dye. This kit contains pHLys Green, which is highly specific to lysosomes and shows pH-dependent changes in fluorescence, and pH-resistant LysoPrime Deep Red. Using these two dyes, lysosomal pH and volume of the same sample can be measured for a detailed analysis of lysosomal function.
|
|||
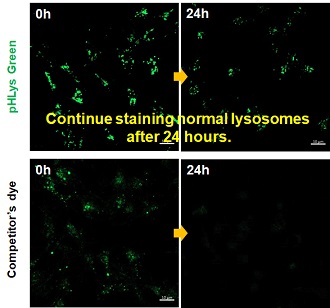
| Existing lysosomal pH detection reagents have issues with dye localization, pH sensitivity, and retention. pHLys Green is a dye that solves these issues. The improved dye retention and localization enable detection of normal lysosomes, and the improved pH sensitivity enables detection of slight pH changes. | |||
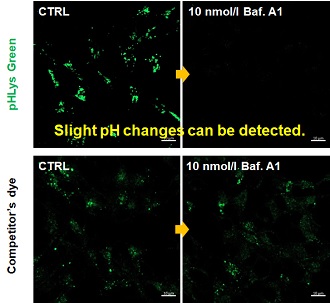
| 1. High sensitive pH detection Comparison of pH response of cells treated with low concentrations of lysosomal acidification inhibitor Bafilomycin A1 |
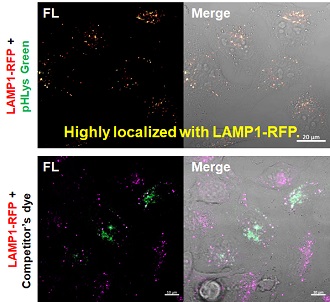
2. High specificity for lysosomes Comparison of specificity for lysosomes using lysosomal marker protein LAMP1-GFP expressing cells |
3. High retention in lysosomes Comparison of intracellular retention |
|
 |
 |
 |
|
|
Product in Use: Related Product: |
|||