|
Recent research on senescence is revealing that senescence is related to lipid droplet and lysosomal dysfunction, which impair cell function. Here are some of the papers showing that these processes leads to neurodegenerative diseases. Cellular senescence, a state of irreversible growth arrest, is closely linked to neurodegeneration through the accumulation of damaged cells in the nervous system. Lipid droplets, which store excess lipids, can accumulate in aging or stressed cells, contributing to cellular dysfunction and exacerbating neurodegenerative processes. Lysosomal dysfunction plays a central role in both lipid accumulation and the removal of cellular waste, and its impairment leads to the accumulation of toxic substances that further drive neurodegeneration. Together, these mechanisms create a cycle of cellular damage that accelerates the progression of neurodegenerative diseases such as Alzheimer's and Parkinson's. |
|||
| Lipid accumulation drives cellular senescence in dopaminergic neurons Click here for the original article: Taylor Russo, et. al., Aging, 2024. |
Senescent glia link mitochondrial dysfunction and lipid accumulation Click here for the original article: China Byrns, et. al., Nature, 2024.
|
BHLHE40/41 regulate microglia and peripheral macrophage responses associated with Alzheimer’s disease and other disorders of lipid-rich tissues Click here for the original article: Anna Podleśny-Drabiniok, et. al., Nature Communications, 2024. |
|
|
Point of Interest - Mutations in the lysosomal enzyme β-glucocerebrosidase cause lipid accumulation that drives cellular senescence in dopaminergic neurons in PD. - Lipid droplet aggregation and lysosomal dysfunction may trigger cellular senescence leading to neurodegeneration in PD. |
Point of Interest - AP1+ senescent glia cause lipid accumulation in non-senescent glia and increase senescence markers. - Targeting AP1 in senescent glia extends lifespan, but increases oxidative damage in the brain and neuronal mitochondrial function remains poor. |
Point of Interest - DLAMs are macrophage subpopulations with similar transcriptional activation states found in aging brains and other diseased lipid-rich tissues. - Targeting BHLHE40/41, transcriptional regulators of DALM, may improve cholesterol clearance and lysosomal function in Alzheimer's disease therapies.
|
|
| Related Techniques | |||
| Cellular senescence detection | SPiDER-βGal for live-cell imaging or flow cytometry / microplate reader / tissue samples NEW SPiDER-βGal Blue for fixed cell and for multiple staining with immunostaining and other methods |
||
| Lipid Droplet detection | Lipid Droplet Assay Kit - Blue / Deep Red | ||
| Lipid Droplet Staining | Lipi-Blue/ Green/ Red/ Deep Red | ||
| Lysosomal function | Lysosomal Acidic pH Detection Kit-Green/Red and Green/Deep Red | ||
| First-time autophagy research | Autophagic Flux Assay Kit | ||
| Mitochondrial membrane potential detection | JC-1 MitoMP Detection Kit, MT-1 MitoMP Detection Kit | ||
| Mitochondrial superoxide detection | MitoBright ROS Deep Red - Mitochondrial Superoxide Detection | ||
| Glycolysis/Oxidative phosphorylation Assay | Glycolysis/OXPHOS Assay Kit | ||
| Glutathione Quantification | GSSG/GSH Quantification Kit | ||
 |
|||
| Related Applications | |||
Co-staining with Lipid droplet and SA-β-Gal in fixed cells |
|||
|
1. A549 (2 x 104) cells were seeded onto µ-slide 8 well plates (ibidi) and cultured overnight in a 37°C CO2 incubator.
|
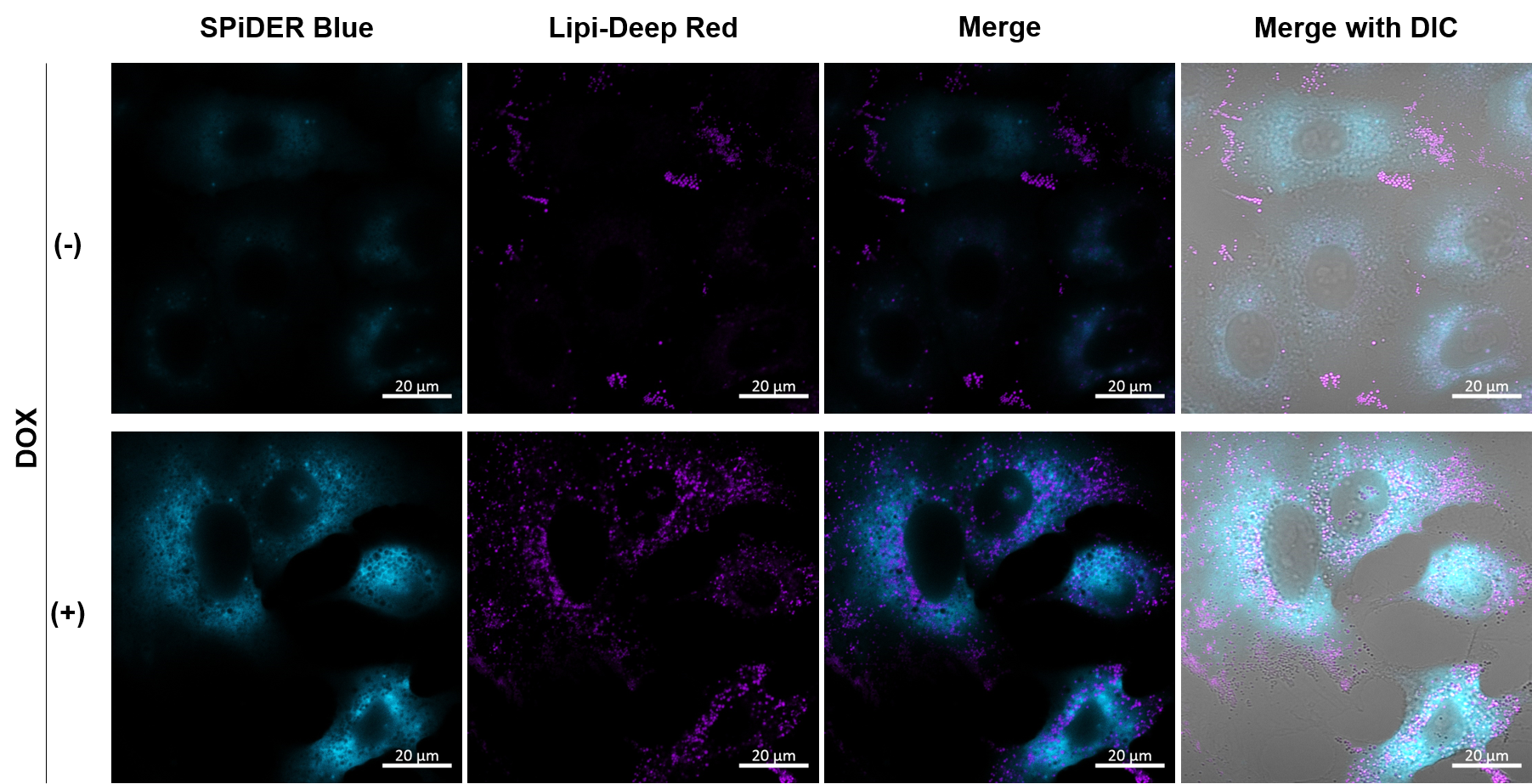
Imaging analysis of lipid droplet accumulation in senescent cells was performed using normal A549 cells (CTRL) and cells induced senescence by Doxorubicin treatment (DOX). SA-β-Gal was detected as a senescence marker with Cellular Senescence Detection Kit - SPiDER Blue, and lipid droplets were detected with Lipi-Deep Red. As a result, the signal of Lipi-Deep Red was increased in SA-β-Gal-positive senescent cells.
[Detection conditions] SPiDE Blue: 405 nm (Ex), 400–550 nm (Em), 1.0%, 600V
|
||
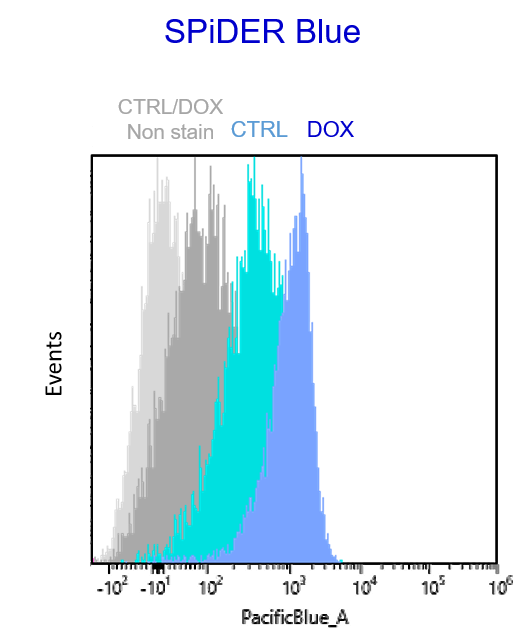
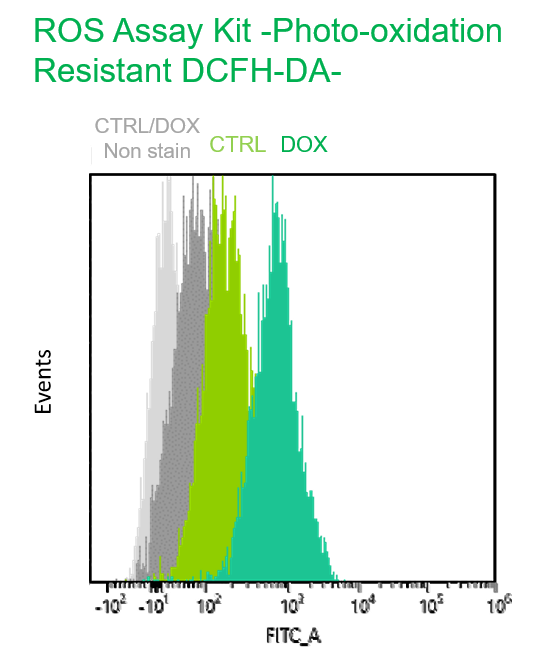
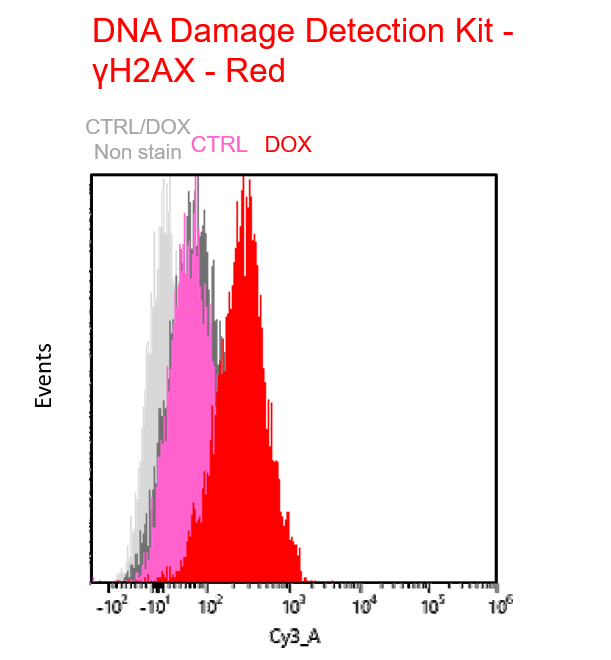
Multiple staining with oxidative stress-related markers using Doxorubicin-induced senescent cells(flow cytometry) |
|||
|
Using A549 cells induced to senescence by doxorubicin (DOX) and normal cells (CTRL), changes in oxidative stress-related markers in senescent cells were analyzed by flow cytometry with multiple staining. SA-βGal as a senescence marker was detected by Cellular Senescence Detection Kit - SPiDER Blue, total ROS as an oxidative stress marker was detected by ROS Assay Kit - Photo-oxidation Resistant DCFH-DA-, and γH2AX as a DNA damage marker was detected by DNA Damage Detection Kit - γH2AX-Red. As a result, total ROS and γH2AX were increased in SA-βGal-positive senescent cells, and the increase in oxidative stress-related markers associated with cellular senescence could be detected by multiple staining. <Experimental Procedure> |
|||