General Information
DNA damage in normal cells may be caused by repeated cell division and oxidative stress. Cellular senescence, a state of irreversible growth arrest, can be triggered in response to DNA-damage. Senescence-associated β-galactosidase (SA-β-gal), which is overexpressed in senescent cells, has been widely used as a marker of cellular senescence 1, 2).
This kit allows for the detection of SA-β-gal with high sensitivity and ease of use. Because SPiDER Blue emits blue fluorescence after reacting with SA-β-gal in fixed cells, it is possible to co-stain with green or red fluorescent probes and fluorescent labeled antibodies for immunostaining.
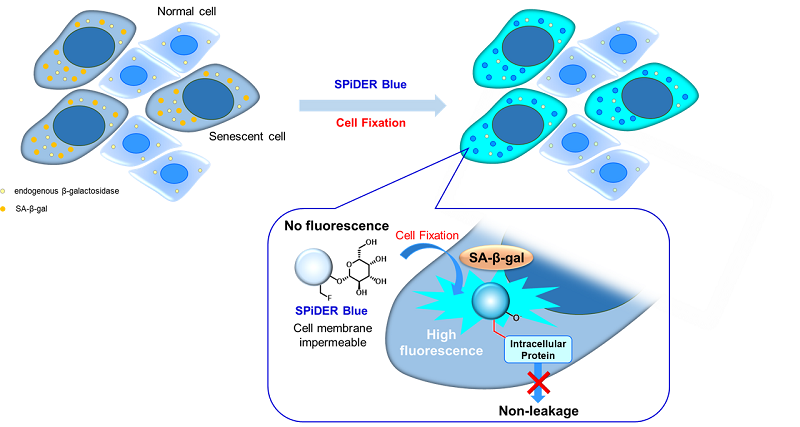
Fig. 1 Detection mechanism of senescent cells by SPiDER Blue
Fluorescent Property
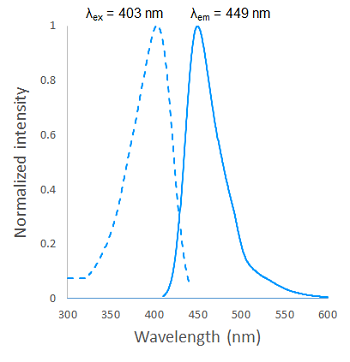
Fig. 2 Excitation and emission spectra of SPiDER Blue after reaction with β-galactosidase
Kit Contents
1 plate (6-well plate)
- SPiDER Blue 10 µl × 1
- Assay Buffer 13 ml × 1
Storage Condition
Store at -20°C.
Required Equipment and Materials
- Micropipettes
- Phosphate buffered saline (PBS)
- Paraformaldehyde (PFA) / PBS solution (4%)
Preparation of Solutions
Preparation of SPiDER Blue working solution
Dilute 20 mmol/l SPiDER Blue with Assay Buffer to prepare a 15 μmol/l SPiDER Blue working solution.
- In order to prepare 2 ml of the SPiDER Blue working solution, dilute 1.5 μl of SPiDER Blue with 2 ml of Assay Buffer.
- The optimal concentration of SPiDER Blue depends on the cells; consider a range of 10-20 μmol/l as a guide.
- Please use the SPiDER Blue working solution on the same day as preparation.
General Protocol
- Seed cells in a dish and culture them at 37°C in an incubator equilibrated with 95% air and 5% CO2.
- Remove the culture medium and wash the cells with PBS.
- Add 4% paraformaldehyde (PFA) / PBS solution to the cells and incubate at room temperature for 30 minutes.
- Discard the 4% PFA / PBS solution and wash the cells with PBS.
- Add 15 µmol/l SPiDER Blue working solution and incubate at 37°C for 30 minutes.
- We do not recommend using a 5% CO2 incubator for fixed cell experiments. If incubation is done in a 5% CO2 incubator, the pH of the buffer may become acidic. Acidic pH results in a higher background from the endogenous β-galactosidase activity and could make it difficult to distinguish between normal cells and senescent cells.
- Remove the working solution, and wash the cells with PBS.
- Add PBS and observe the cells under a fluorescence microscope.
Usage Example 1
Fluorescence imaging of SA-β-gal in doxorubicin-treated A549 cells
- A549 cells (1 × 106 cells/dish, DMEM containing 10% fetal bovine serum and 1% penicillin-streptomycin) were seeded on a 10-cm dish and cultured at 37°C overnight in a 5% CO2 incubator.
- The medium was removed and the cells were washed with serum-free DMEM once. Doxorubicin (DOX) solution (0.2 μmol/l in serum-free DMEM) was added, and the cells were cultured at 37°C for 3 days in a 5% CO2 incubator.
- The DOX solution was removed, and the cells were washed with serum-free DMEM once. DMEM containing 10% fetal bovine serum and 1% penicillin-streptomycin was added, and the cells were cultured at 37°C for 3 days in a 5% CO2 incubator.
- The trypsinized DOX-treated and untreated A549 cell suspension (1 × 105 cells/ml, DMEM containing 10% fetal bovine serum and 1% penicillin-streptomycin) were seeded (200 μl/well) on a μ-slide 8-well plate (ibidi) and cultured at 37°C overnight in a 5% CO2 incubator.
- The medium was removed, and the cells were washed with PBS once. A 4% paraformaldehyde (PFA) / PBS solution was added to the cells and incubated at room temperature for 30 minutes.
- The 4% PFA / PBS solution was removed, and the cells were washed with PBS once.
- SPiDER Blue working solution (15 µmol/l in Assay Buffer) was added to the cells and incubated at 37°C for 30 minutes.
- The working solution was removed, and the cells were washed with PBS once. PBS was added, and the cells were observed by confocal fluorescence microscopy (40×).
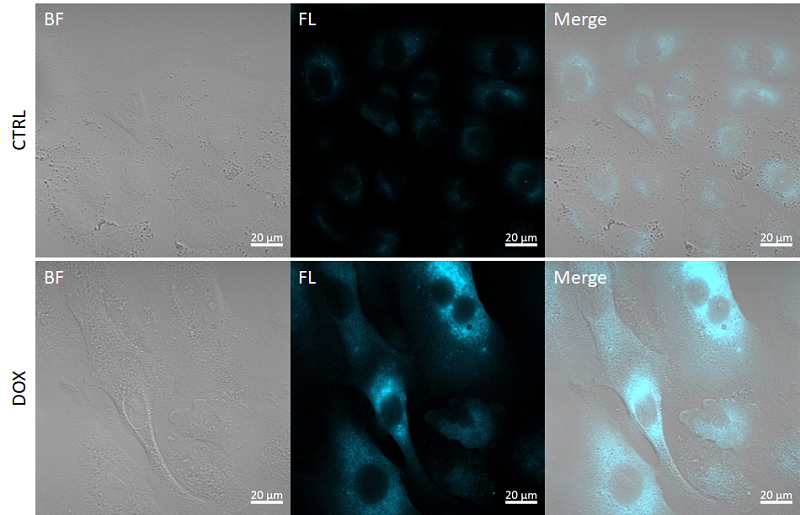
Fig. 3 Fluorescence imaging of SA-β-gal
CTRL: Normal condition, DOX.: Senescent condition
Filter sets: 405 nm (Ex), 400–500 nm (Em)
Usage Example 2
Fluorescence imaging of SA-β-gal in non-senescent and senescent WI-38 cells with different passage numbers.
- WI-38 cells (1 × 106 cells/dish, MEM containing 10% fetal bovine serum and 1% penicillin-streptomycin) of passage number 4 and 12 were seeded in 10-cm dishes and cultured at 37°C overnight in a 5% CO2 incubator.
- The trypsinized WI-38 cell suspension (1 × 105 cells/ml, MEM containing 10% fetal bovine serum and 1% penicillin/streptomycin) were seeded (200 μl/well) on a μ-slide 8-well plate (ibidi) and cultured at 37°C overnight in a 5% CO2 incubator.
- The medium was removed, and the cells were washed with PBS once. A 4% paraformaldehyde (PFA) / PBS solution was added to the cells and incubated at room temperature for 30 minutes.
- The 4% PFA / PBS solution was removed, and the cells were washed with PBS once.
- SPiDER Blue working solution (15 µmol/l in Assay Buffer) was added to the cells and incubated at 37°C for 30 minutes.
- The working solution was removed, and the cells were washed with PBS once. PBS was added, and the cells were observed by confocal fluorescence microscopy (40×).
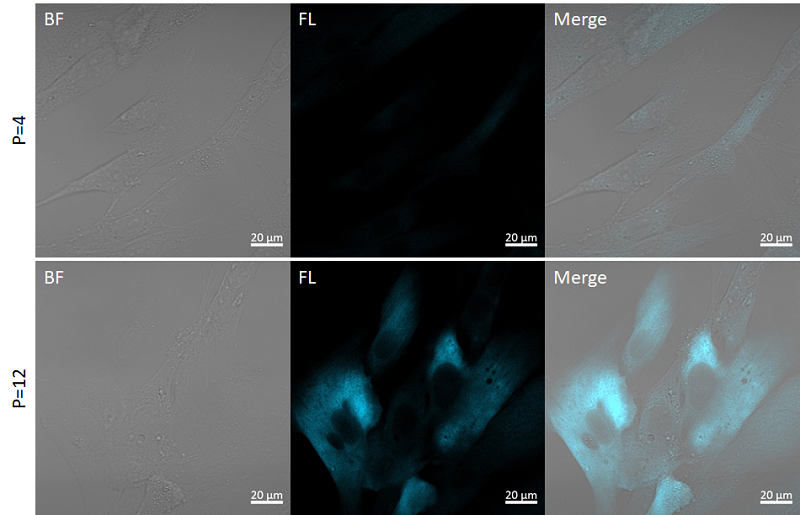
Fig. 4 Fluorescence imaging of SA-β-gal
P=4: Passage number 4, P=12: Passage number 12
Filter sets: 405 nm (Ex), 400–500 nm (Em)
References
- Dimri, G. P. et al., Cell Biology, 1995, 92, 9363–9367.
- Park, A. M. et al., J. Biol. Chem., 2018, 293(41), 15815-15826.
Frequently Asked Questions / Reference
SG07: Cellular Senescence Detection Kit - SPiDER Blue
Revised Dec., 12, 2024


 Hidden sections will not be printed.
Hidden sections will not be printed.

