General Infomation
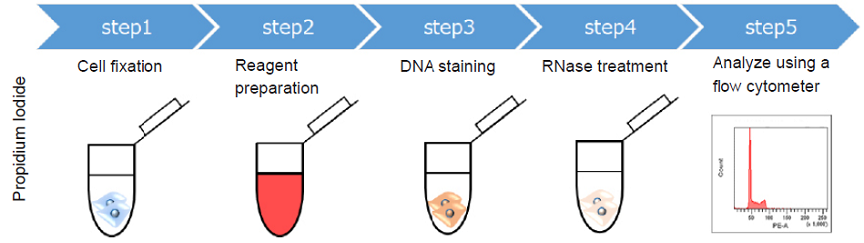
Cell cycle is associated with the most important phenomena of cell growth. Various phases of the cell cycle can be analyzed by staining DNA and using a flow cytometer. Propidium Iodide (PI) is a widely used dye for cell cycle analysis using a flow cytometer. However, PI has a number of limitations because of the following reasons.
- Cell fixation is required (non-cell membrane permeability)
- RNase treatment is needed (PI is non-selective toward DNA)
- Necessity of compensation (PI is detected by the PE channel and when using antibody conjugated dye detected FITC, compensation is necessary).
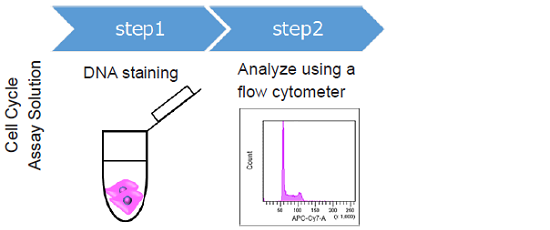
The Cell Cycle Assay Solution overcomes these limitations. This solution offers a much simpler procedure when compared with that using PI. Two different color options (Deep Red and Blue) are provided as ready-to-use solutions. Furtheremore, either solution enables the determination of the ratio of the G0/G1 phase, S phase, and G2/M phase using a flow cytometer. The color of either solution can be combined with your experiments.
Contents
| 50 tests | ||
| C548 | Cell Cycle Assay Solution Deep Red | 250 µl x 1 |
| C549 | Cell Cycle Assay Solution Blue | 250 µl x 1 |
- C548 and C549 are provided as aqueous solutions.
Storage Condition
C548 : Store at -20°C and protected from light.
C549 : Store at -20°C.
Required Equipment and Materials
- Flow cytometer
- Cell strainer for flow cytometer
- Incubator (37°C)
- Micropipettes (100–1000 µl, 1–20 µl)
- Conical tubes - Microcentrifuge tubes
- Phosphate buffered saline (PBS)
Preparation
Allow the Cell Cycle Assay Solution to equilibrate at room temperature before use.
- Centrifuge the tube briefly before opening the cap because the content may be on the tube wall or on the ceiling of the cap.
Precaution
Optimize the gating (FSC, SSC) after the staining because the scattergram on FSC and SSC are changed by the staining process with the Cell Cycle Assay Solution.
- Refer Figure 5
Remaining trypsin may result obtaining improper histogram in Flow cytometric analysis. Carefully remove trypsin by washing before staining.
- Refer Figure 6.
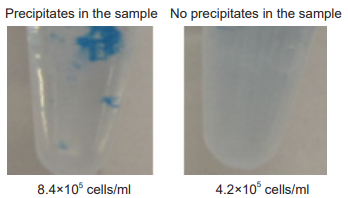
Too much number of cells may cause precipitation in the sample. Optimization of cell number is required.
- Refer Figure 7.
General protocol
For non-fixed cells
- Prepare a cell suspension (1–5 x 105 cells/ml) in a 1.5 ml microcentrifuge tube.
- Centrifuge at 300 x g for 5 minutes and discard the supernatant.
- Add 500 µl of PBS to each microcentrifuge tube, suspend by pipetting, centrifuge at 300 × g for 5 minutes and discard the supernatant.
- Add 500 µl of PBS to each microcentrifuge tube and add 5 µl of Cell Cycle Assay Solution.
- Incubate at 37℃ for 15 minutes and protect from light.
- Pass through a cell strainer and analyze samples using a flow cytometer.
- *Recommended filter settings are shown in Table 1.
For fixed cells
- Prepare a cell suspension (1–5 x 105 cells/ml) in a 1.5 ml microcentrifuge tube.
- Centrifuge at 300 x g for 5 minutes and discard the supernatant.
- Fix the cells by using one of the following protocols or your original fixation protocol.
- Add 5 µl of C548 or C549 and incubate at 37℃ for 15 minutes, protect from light.
- Pass through a cell strainer and analyze samples using a flow cytometer.
- Recommended filter settings are shown in Table 1.
Using a 4% paraformaldehyde (PFA) /PBS solution.
- Add 1 ml of a 4% PFA/PBS solution to a microcentrifuge tube and incubate at room temperature for 20 minutes.
- Centrifuge at 300 x g for 5 minutes and discard the supernatant.
- Add 500 µl of PBS to the microcentrifuge tube, suspend by pipetting, centrifuge at 300 x g for 5 minutes. Discard the supernatant and add 500 µl of PBS to each microcentrifuge tube.
Using cold 70% ethanol.
- Add 1 ml of a 70% ethanol to a microcentrifuge tube and incubate at -20℃ for 120 minutes.
- Centrifuge at 300 x for 5 minutes and discard the supernatant.
- Add 500 µl of PBS to the microcentrifuge tube, suspend by pipetting, centrifuge at 300 x g for 5 minutes. Discard the supernatant and add 500 µl of PBS to each microcentrifuge tube.
|
* LP : Long pass filter * BP : Band pass filter |
Usage examples
Cell Cycle Analysis using HeLa cells
- HeLa cells in MEM (containing 10% fetal bovine serum, 1% penicillin-streptmycin) were seeded on a 6-well plate (1–5×105 cells/ml) and cultured overnight in a 37℃ incubator equilibrated with 95% air and 5% CO2.
- After trypsinization, 1 ml of the cell suspension was transferred to a microcentrifuge tube.
- The cells suspension was centrifuged at 3000 x g for 5 minutes.
- The supernatant was discarded and 500 µl PBS was added to the microcentrifuge tube and suspended by pipetting.
- The cells suspension was centrifuged at 300 x g for 5 minutes and the supernatant was discarded.
- The cells were re-suspended in PBS (500 µl) and 5 µl of C548 or C549 was added.
- Each tube was incubated at 37℃ for 15 minutes, protected from light.
- The stained cells were passed through a cell strainer and analyze samples using a flow cytometer.
-
Cell Cycle Assay Solution Deep Red
Excitation: 640 nm
Emission: 780/60 nm
-
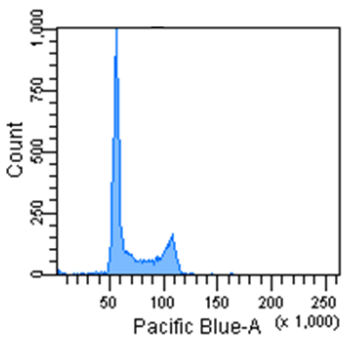
Cell Cycle Assay Solution Blue
Excitation: 405 nm
Emission: 450/50 nm
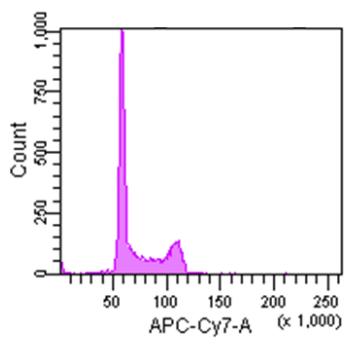
Figure 2 Cell cycle analysis using HeLa cells stained with the Cell Cycle Assay Solution.
Cell Cycle Analysis using Jurkat cells
- Jurkat cells in MEM (containing 10% fetal bovine serum, 1% penicillin-streptmycin) were seeded on a 6-well plate (1–5×105 cells/ml) and cultured overnight in a 37℃ incubator equilibrated with 95% air and 5% CO2.
- After trypsinization, 1 ml of the cell suspension was transferred to a microcentrifuge tube.
- The cells suspension was centrifuged at 300× g for 5 minutes.
- The supernatant was discarded and 500 µl PBS was added to the microcentrifuge tube and suspended by pipetting.
- The cells suspension was centrifuged at 300× g for 5 minutes and the supernatant was discarded.
- The cells were re-suspended in PBS (500 µl) and 5 µl of C548 or C549 was added.
- Each tube was incubated at 37℃ for 15 minutes, protected from light.
- The stained cells were passed through a cell strainer and analyze samples using a flow cytometer.
-
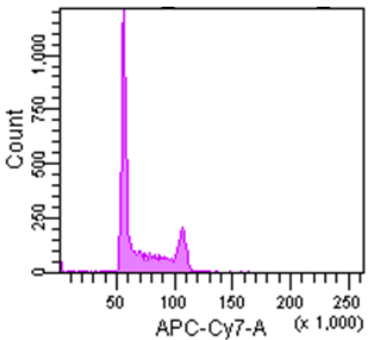
Cell Cycle Assay Solution Deep Red
Excitation: 640 nm
Emission: 780/60 nm
-
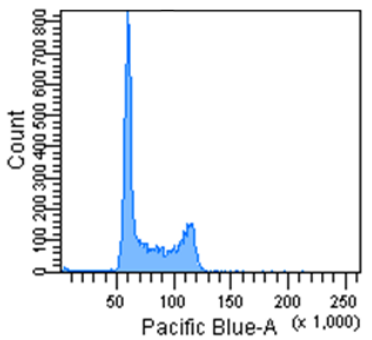
Cell Cycle Assay Solution Blue
Excitation: 405 nm
Emission: 450/50 nm
Figure 3 Cell cycle analysis using Jurkat cells stained with the Cell Cycle Assay Solution.
Troubleshooting
Optimize the gating (FSC, SSC) after staining because the FSC and SSC signals are changed by the staining.
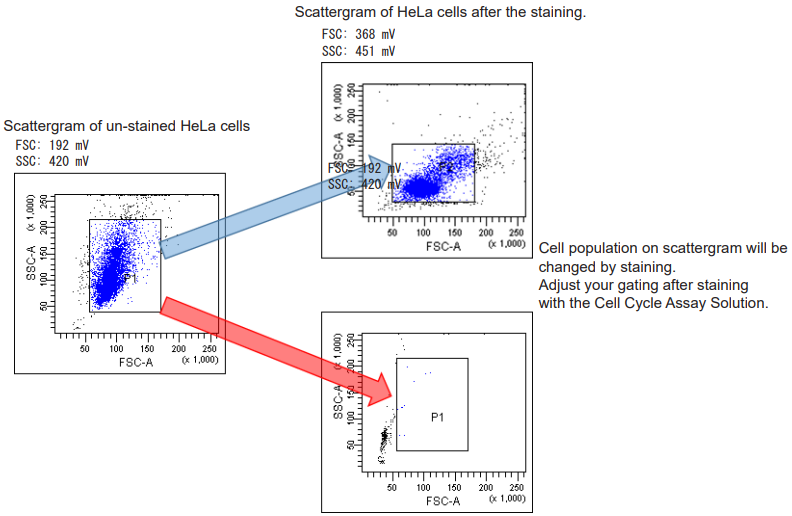
Figure 5 Scattergram of HeLa cells before or after staining.
Negative effect of trypsin
Carefully remove trypsin by washing. Remaing trypsin may result obtaining improper histogram as follows.
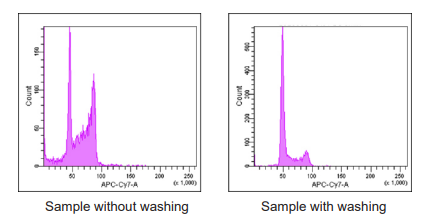
Figure 6 Histogram of HeLa cells in the presence/absence of trypsin.
Precipitates in the sample
When precipitates are observed, reduce the number of cell in the sample.
-
cell line HeLa cell number 4.2–8.4 ×105 cells/ml amount of reagent 5 µl incubation 37℃ , 15 minutes
Figure.7 Precipitates in the sample of HeLa cells.
C548_C549: Cell Cycle Assay Solution Deep Red/Blue
Revised Apr., 11, 2024


 Hidden sections will not be printed.
Hidden sections will not be printed.