Autophagy : Reagent Selection Guide
Topics
- What is Autophagy?
- Autophagy Reagents Selection Guide
- Principle of Autophagy Reagents
- Typical Examples of Article in Use
- Experimental Example: Analysis of autolysosome formation inhibition using Bafilomycin A1 by confocal fluorescence microscopy
- Experimental Example: Analysis of lysosomal protein degradation inhibition using E64d/Pepstatin A by confocal fluorescence microscopy
- Experimental Example: Time-lapse imaging of autophagy with DALGreen
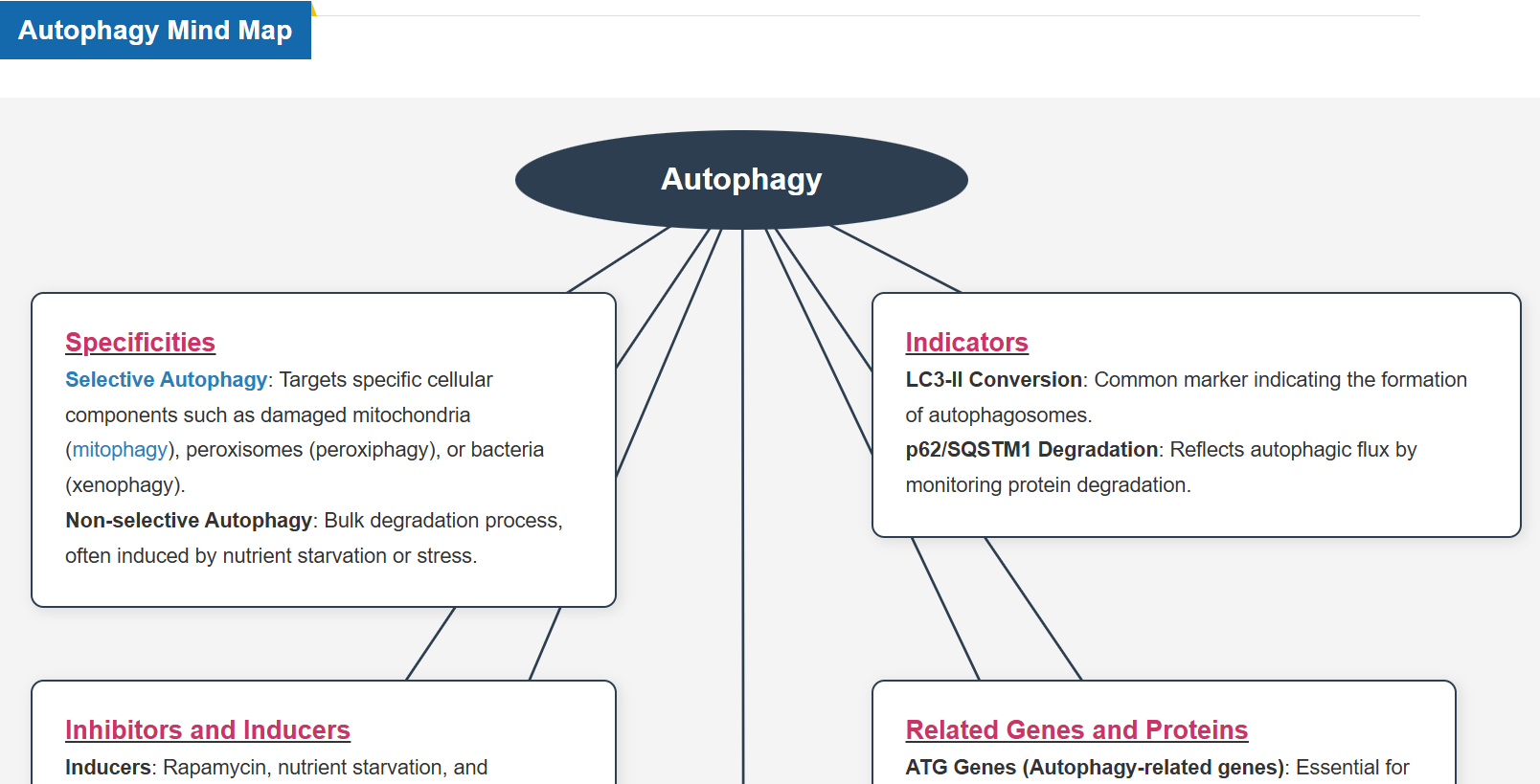
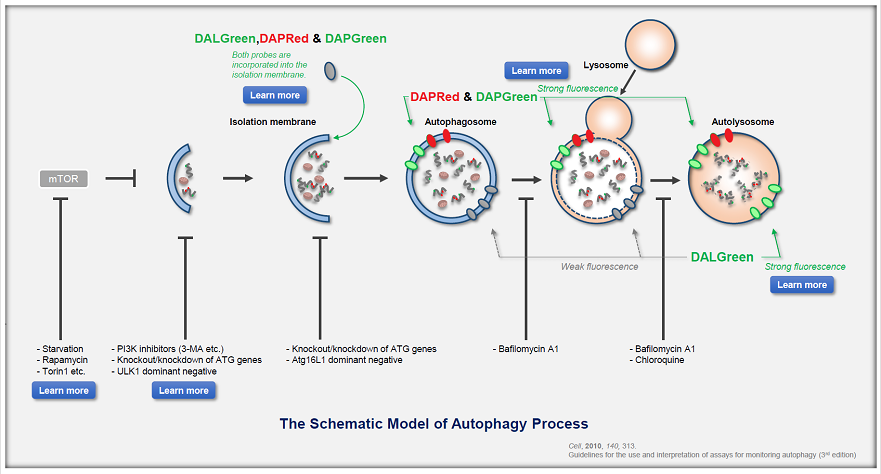
What is Autophagy?
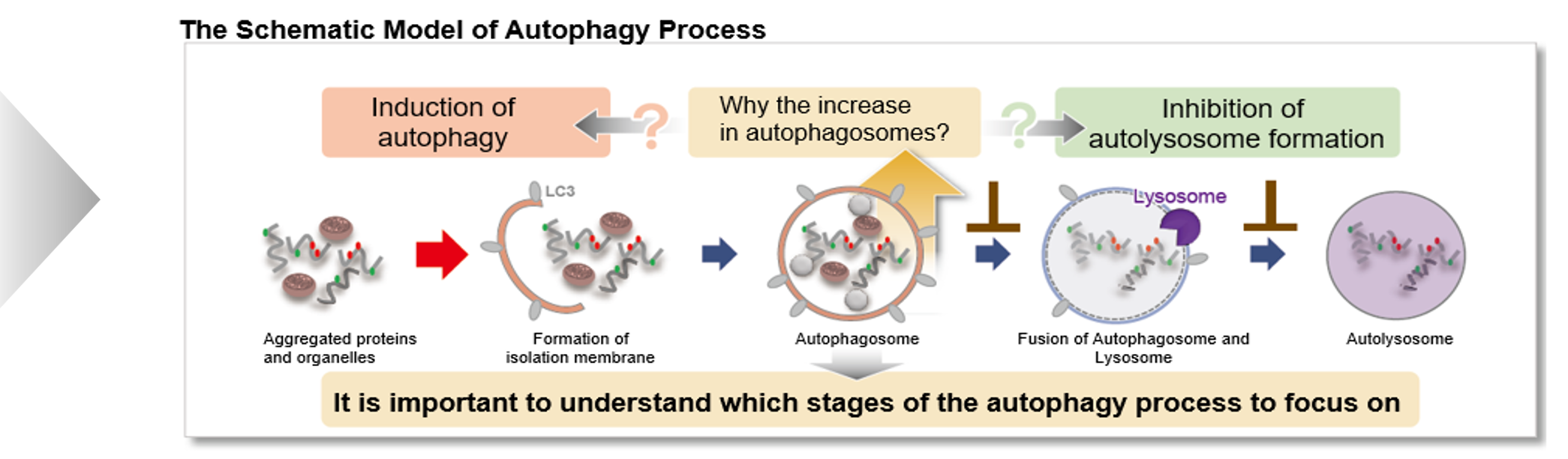
Autophagy is a degradation process of cytoplasmic dysfunctional proteins and organelles. In this process, an isolation membrane composed of a double membrane appears in the cytosol, gradually expands, encloses the aggregated proteins and damaged organelles, and closes to form autophagosomes. The autophagosomes are fused with lysosomes to form autolysosomes, which have an acidic environment. The contents in autolysosomes are decomposed by digestive enzymes in lysosomes. Since this cellular function is said to be related to aging as well as neurodegenerative diseases such as Parkinson’s disease, a simple autophagy detection method is being required.
Autophagy Reagents Selection Guide
Depending on the method and purpose of autophagy assessment, three types of fluorescent small molecule reagents and kits are available.
Select by targets
| Small Fluorescent Molecules | Fluorescent Protein | ||||||
|---|---|---|---|---|---|---|---|
| DAPGreen | DAPRed | DALGreen | Autophagic Flux Assay Kit | GFP-LC3 | RFP-LC3 | mRFP-GFP-LC3 | |
| Autophagosome | ✓ | ✓ | - | Autophagosomes and autolysosomes are each detectable. | ✓ | ✓ | Autophagosomes and autolysosomes are each detectable. |
| Autolysosome | ✓ | ✓ | ✓ | - | ✓ | ||
| Transfection | No need for transfection | ✓ | |||||
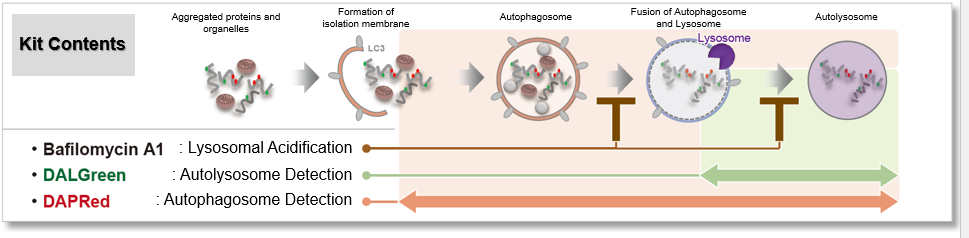
Autophagic Flux Assay Kit: This kit contains autophagosome and autolysosome detection dye (DAPRed), autolysosome detection dye (DALGreen), and lysosomal acidification inhibitor (bafilomycin A1). The Autophagic Flux Assay Kit allows the accurate evaluation of autophagic flux by monitoring autophagosome formation, lysosome fusion, and digestion of contents.
Select by detector and fluorescence property
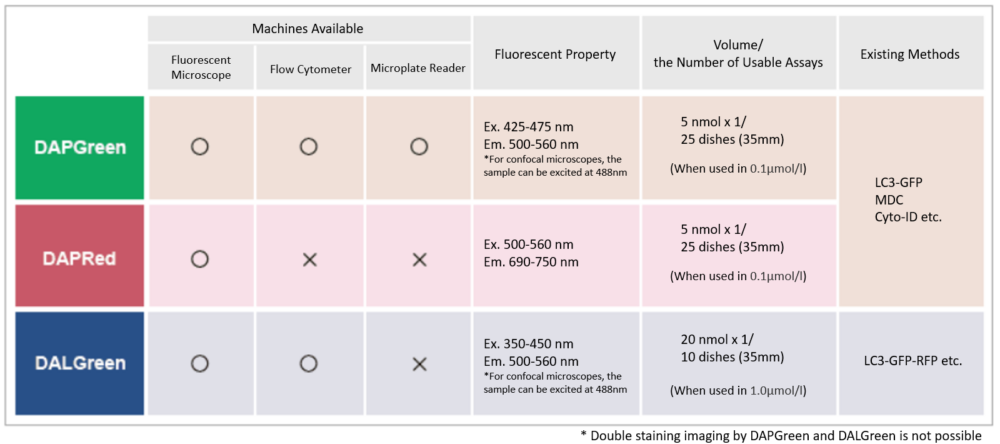
There are several autophagy-related products available, which one to choose?
|
Example: Screen the autophagy activity of a drug
|
Principle of Autophagy Reagents
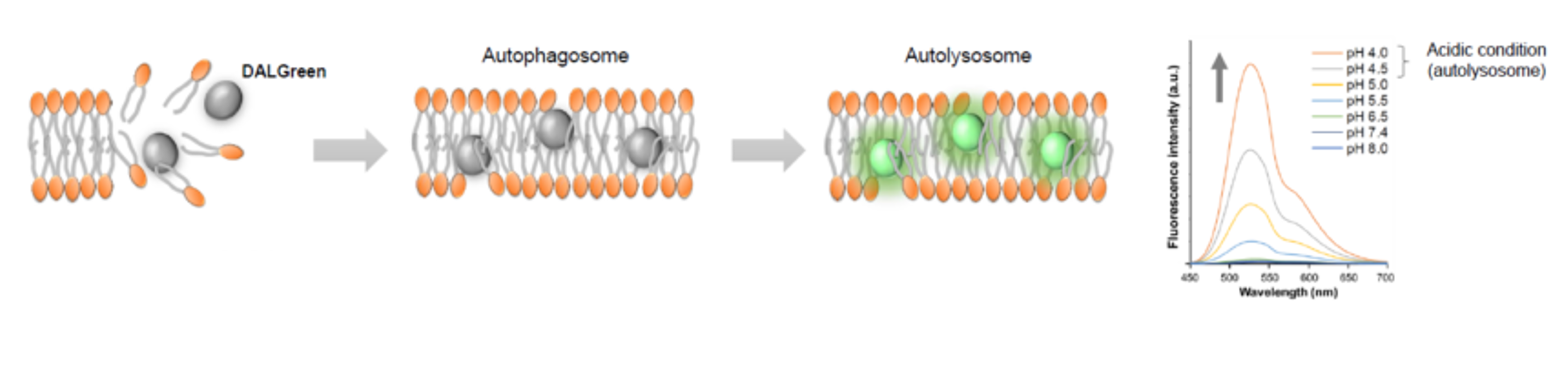
DALGreen / DAPGreen is incorporated into hydrophobic lipid bilayers due to its similar structure to membrane phospholipids such as phosphatidylethanolamine.
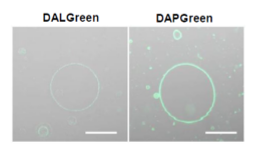
DALGreen and DAPGreen were each incorporated during liposome membrane formation and observed by confocal microscopy; DAPGreen showed strong fluorescence upon incorporation into the lipid bilayer, whereas weak fluorescence was observed for DALGreen. Scale bar: 20 μm
DAPGreen: fluorescence intensity increases in response to the hydrophobic field environment within the lipid bilayer (detects both autophagosomes and autolysosomes)
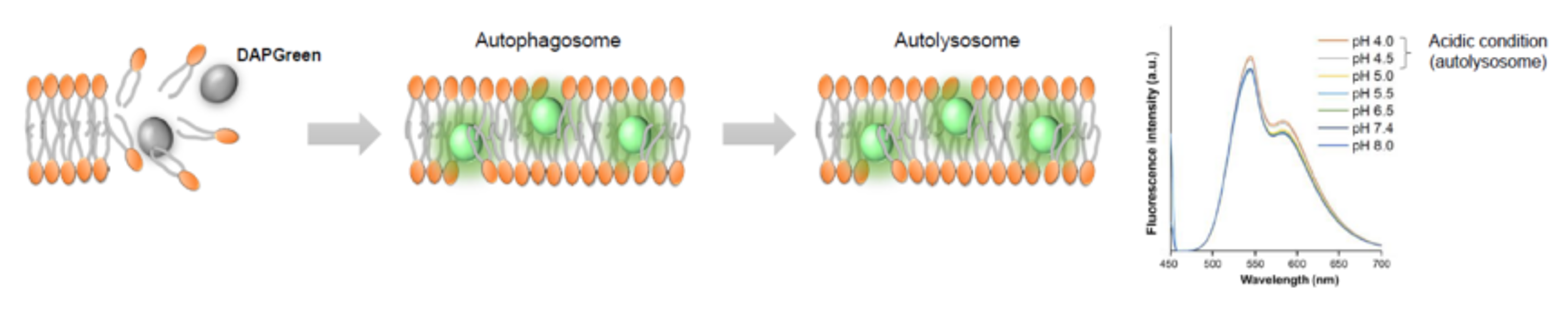
DALGreen : incorporated into lipid bilayers, fluorescence intensity increases in acidic environment (detects autolysosomes)
For detailed experimental results, including the specificity of autophagy, see the original paper here.
DAPGreen/DALGreen: H. Iwashita, et al., "Small fluorescent molecules for monitoring autophagic flux", FEBS Letters., 2018, 592, (4), 559–567.
DAPRed:H. Sakurai, et al, "Development of small fluorescent probes for the analysis of autophagy kinetics, iScience, 2023, 26, 107218.
Typical Examples of Article in Use
| Title | Discovery and Structure-Based Optimization of Novel Atg4B Inhibitors for the Treatment of Castration-Resistant Prostate Cancer Kudo, Y. et al., Journal of Medical Chemistry, 2022, 65(6) |
| Purpose | Evaluation of ATG4B inhibitors using autophagy activity as an indicator. |
|
Product/ |
DAPGreen/Microscope |
| Title | S-Nitrosylation of p62 Inhibits Autophagic Flux to Promote α-Synuclein Secretion and Spread in Parkinson's Disease and Lewy Body Dementia Oh, C. et al., Journal of Neuroscience, 2022, 41(14), 3011-3024 |
| Purpose | Confirmation that p62(C331A) knock-in mutant causes autophagy inhibition (decrease in autolysosomes). |
| Product/ Method |
DALGreen&DAPRed (Autophagic Flux Assay Kit)/Microscope |
| Title | Scd1 and monounsaturated lipids are required for autophagy and survival of adipocytes Mori, H. et al., Molecular Metabolism, 2024, 83, 101916 |
| Purpose | Confirmation that SCD1KO causes autophagy inhibition (decrease in autolysosomes). |
| Product/ Method |
DALGreen&DAPRed (Autophagic Flux Assay Kit)/Microscope |
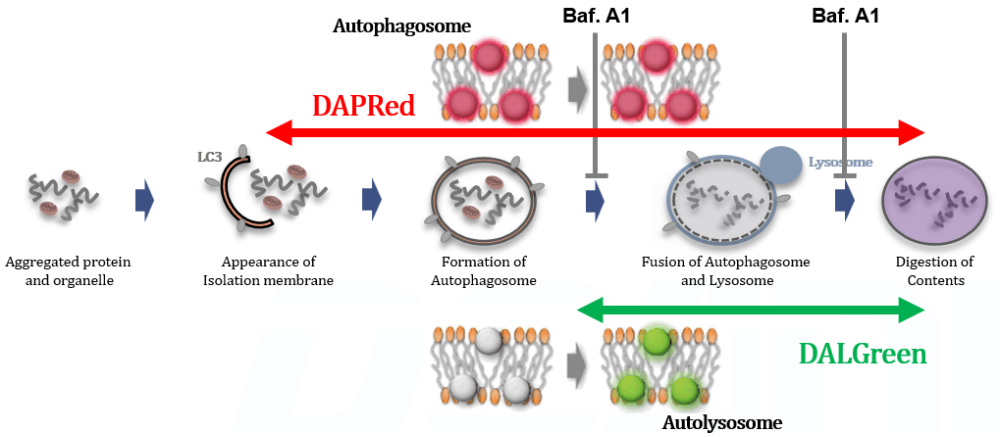
Experimental Example: Analysis of autolysosome formation inhibition using Bafilomycin A1 by confocal fluorescence microscopy
DALGreen and DAPRed labeled HeLa cells were used to evaluate changes in autophagic flux induced by the lysosomal acidification inhibitor bafilomycin A1 (Baf. A1). Compared to starvation conditions, the fluorescence signals of DALGreen were decreased under inhibited conditions of autolysosome formation by the addition of Baf. A1. In contrast, the fluorescence signals of DAPRed were increased under the same conditions, indicating that Baf. A1 led to the accumulation of autophagosome.
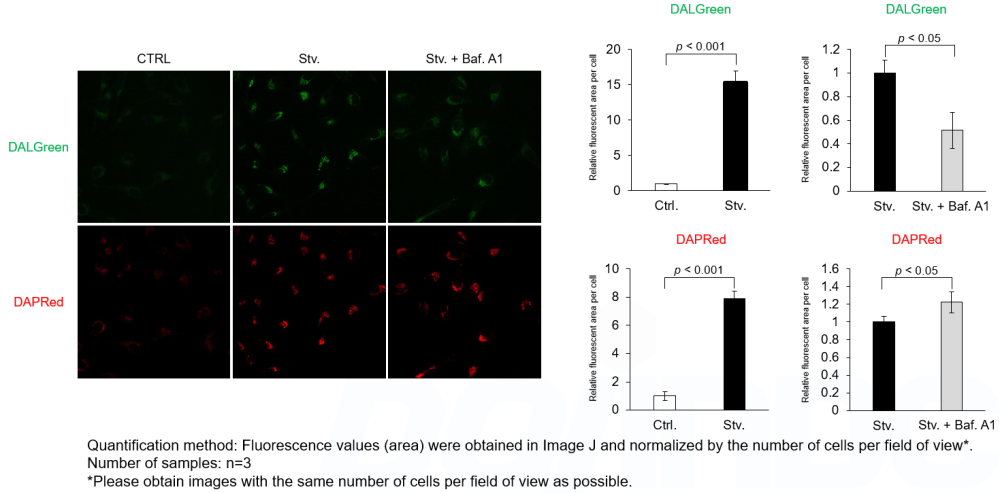
<Experimental Conditions>
CTRL: Normal condition, Stv.: Induction of autophagy, Stv. + Baf. A1: Inhibition of autolysosome formation
DALGreen filter set: 488 nm (Ex), 490–550 nm (Em)
DAPRed filter set: 561 nm (Ex), 565–700 nm (Em)
<Procedure>
1. HeLa cells were seeded (1.0 x 104 cells/well) on a μ-slide 8 well plate (ibidi) and cultured overnight at 37°C in an incubator equilibrated with 95% air and 5% CO2.
2. After washing twice with MEM containing 10% fetal bovine serum, 200 μl of DALGreen/DAPRed working solution (DALGreen: 1 µmol/l, DAPRed: 0.2 µmol/l) and the cells were incubated at 37°C for 30 minutes.
3. The supernatant was discarded, and the cells were washed twice with MEM containing 10% fetal bovine serum.
4. Samples were prepared under the following conditions.
• MEM containing 10% fetal bovine serum (200 µl) was added to the well, and the cells were incubated at 37 °C for 2 hours 20 minutes. (Control)
• Amino acid-free medium (FUJIFILM Wako Pure Chemical Industries, Ltd., Catalogue code: 048-33575) (200 μl) was added to the well, and the cells were incubated at 37°C for 2 hours 20 minutes. (Starvation)
• Amino acid-free medium (200 μl) was added to the well, and the cells were incubated at 37°C for 2 hours. The supernatant was discarded, bafilomycin A1 working solution (10,000 times dilution, 200 μl), an inhibitor of lysosomal acidification, was added to the well, and the cells were incubated at 37°C for 20 minutes. (Inhibition of autolysosome formation)
5. The stained cells were observed under a confocal fluorescence microscope.
Experimental Example: Analysis of lysosomal protein degradation inhibition using E64d/Pepstatin A by confocal fluorescence microscopy
DALGreen and DAPRed labeled HeLa cells were used to evaluate changes in autophagic flux induced by the inhibitor of lysosome enzymes E64d/Pepstatin A (Pep A). Compared to starvation conditions, the fluorescence signals of DALGreen and DAPRed were increased due to autolysosome accumulation by the addition of E64d/Pep A.
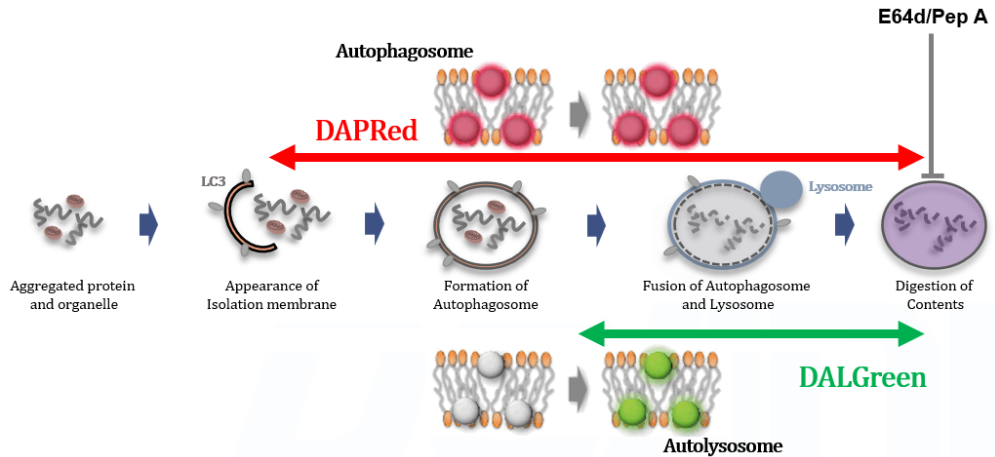
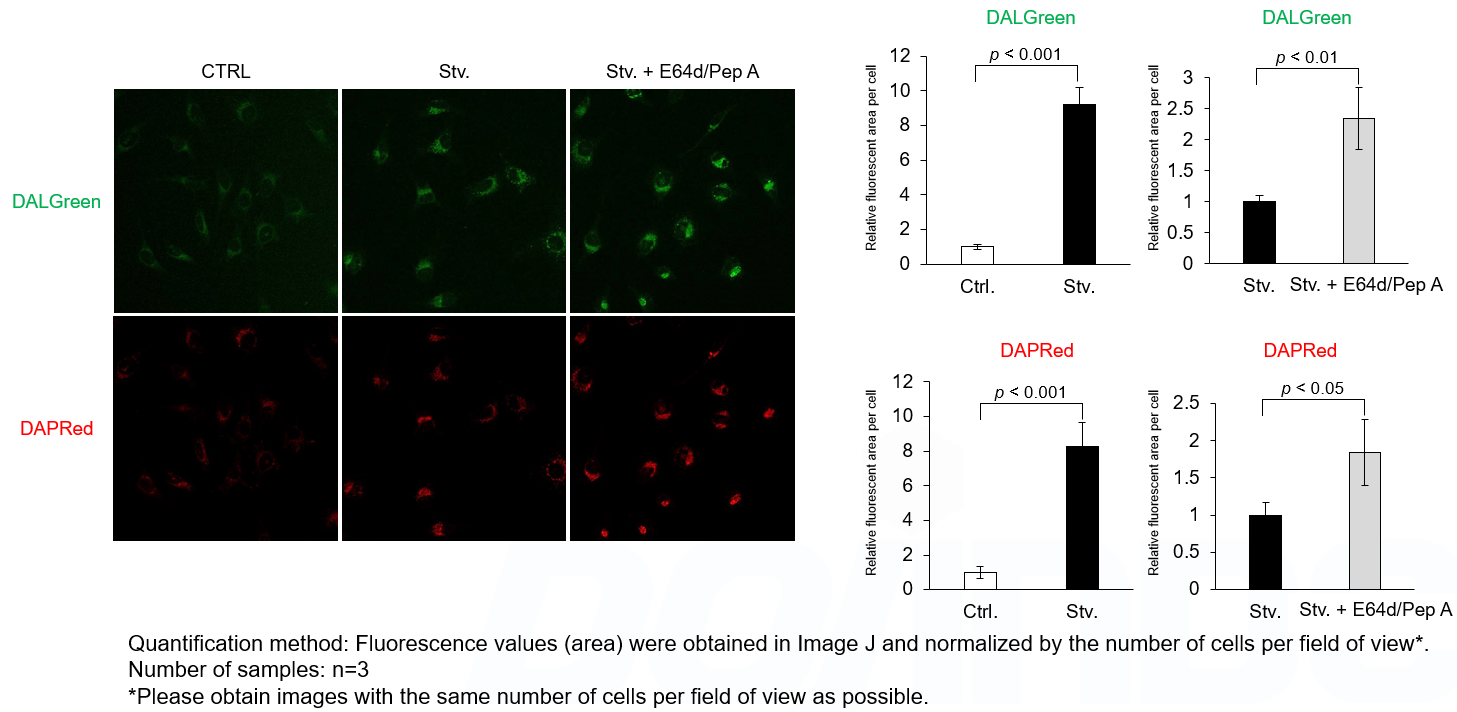
<Experimental Conditions>
CTRL: Normal condition, Stv.: Induction of autophagy, Stv. + E64d/Pep A: Inhibition of lysosomal protein degradation
DALGreen filter set: 488 nm (Ex), 490–550 nm (Em)
DAPRed filter set: 561 nm (Ex), 565–700 nm (Em)
<Procedure>
1. HeLa cells were seeded (1.0 x 104 cells/well) on a μ-slide 8 well plate (ibidi) and cultured overnight at 37 °C in an incubator equilibrated with 95% air and 5% CO2.
2. After washing twice with MEM containing 10% fetal bovine serum, 200 μl of DALGreen/DAPRed working solution (DALGreen: 1 µmol/l, DAPRed: 0.2 µmol/l) and the cells were incubated at 37°C for 30 minutes.
3. The supernatant was discarded, and the cells were washed twice with MEM containing 10% fetal bovine serum.
4. Samples were prepared under the following conditions.
• MEM containing 10% fetal bovine serum (200 µl) was added to the well, and the cells were incubated at 37°C for 2 hours. (Control)
• Amino acid-free medium (FUJIFILM Wako Pure Chemical Industries, Ltd., Catalogue code: 048-33575) (200 μl) was added to the well, and the cells were incubated at 37°C for 2 hours. (Starvation)
• Amino acid-free medium containing E64d/Pep A (10 µg/ml each, 200 μl), an inhibitor of lysosomal protease, was added to the well, and the cells were incubated at 37°C for 2 hours. (Inhibition of lysosomal protein degradation)
5. The stained cells were observed under a confocal fluorescence microscope.
Experimental Example: Time-lapse imaging of autophagy with DALGreen
For more details, click application data
H. Iwashita, H. T. Sakurai, N. Nagahora, M. Ishiyama, K. Shioji, K. Sasamoto, K. Okuma, S. Shimizu, and Y. Ueno, “Small fluorescent molecules for monitoring autophagic flux“, FEBS Lett., 2018, 592, (4), 559–567.